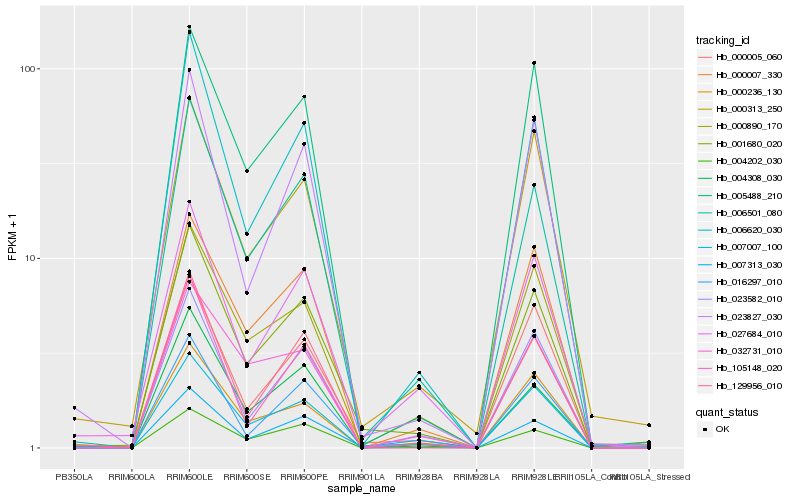
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001680_020 |
0.0 |
desease resistance |
Gene Name: RPW8 |
Disease resistance protein ADR1, putative [Ricinus communis] |
| 2 |
Hb_006501_080 |
0.0704496142 |
- |
- |
PREDICTED: uncharacterized protein LOC105637775 [Jatropha curcas] |
| 3 |
Hb_006620_030 |
0.0880743924 |
- |
- |
nitrate reductase, putative [Ricinus communis] |
| 4 |
Hb_016297_010 |
0.0932619778 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 5 |
Hb_007007_100 |
0.101611171 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 6 |
Hb_000890_170 |
0.1035778773 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 7 |
Hb_027684_010 |
0.1047713082 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 8 |
Hb_000005_060 |
0.1074354784 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 9 |
Hb_004308_030 |
0.1119793795 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 10 |
Hb_023582_010 |
0.1120400213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005488_210 |
0.113871067 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 12 |
Hb_000313_250 |
0.1143120189 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 13 |
Hb_004202_030 |
0.1152995157 |
- |
- |
PREDICTED: uncharacterized protein LOC103449702 [Malus domestica] |
| 14 |
Hb_023827_030 |
0.1153075683 |
- |
- |
PREDICTED: thylakoid lumenal 16.5 kDa protein, chloroplastic-like [Populus euphratica] |
| 15 |
Hb_000236_130 |
0.1155294597 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 16 |
Hb_000007_330 |
0.1160537197 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 17 |
Hb_105148_020 |
0.1164502146 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 18 |
Hb_129956_010 |
0.1204260162 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK2 [Eucalyptus grandis] |
| 19 |
Hb_032731_010 |
0.121166102 |
- |
- |
RING-H2 finger protein ATL5M precursor, putative [Ricinus communis] |
| 20 |
Hb_007313_030 |
0.1213759631 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |