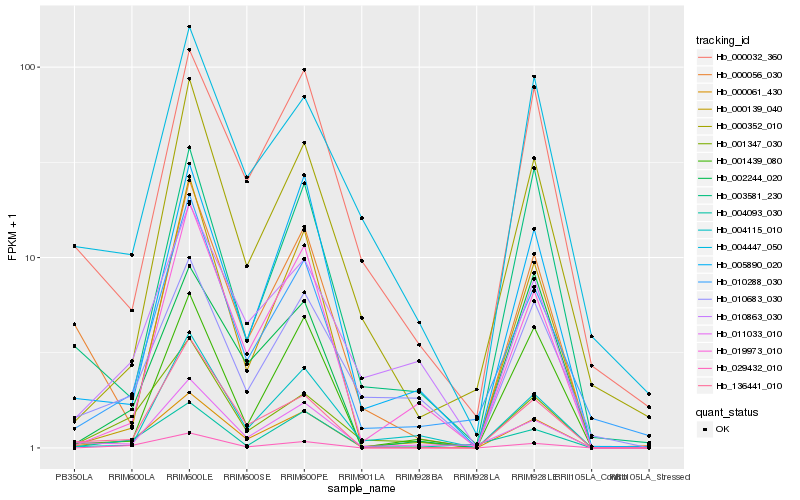
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_430 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011033_010 |
0.1276085727 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 3 |
Hb_136441_010 |
0.1357096277 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 4 |
Hb_005890_020 |
0.1359783287 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 5 |
Hb_001347_030 |
0.1462859299 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_019973_010 |
0.1468966983 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 7 |
Hb_010288_030 |
0.1473438056 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 8 |
Hb_029432_010 |
0.1486521739 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_001439_080 |
0.1509794582 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 10 |
Hb_000139_040 |
0.1531398256 |
- |
- |
spermine synthase, putative [Ricinus communis] |
| 11 |
Hb_000032_360 |
0.1567151767 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit S, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002244_020 |
0.1580215503 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 13 |
Hb_003581_230 |
0.1605050668 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 14 |
Hb_000352_010 |
0.1612457188 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000056_030 |
0.1633824847 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g14390 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010683_030 |
0.1658124421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004093_030 |
0.1671010704 |
- |
- |
hypothetical protein RCOM_1406750 [Ricinus communis] |
| 18 |
Hb_004115_010 |
0.1676730615 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 19 |
Hb_004447_050 |
0.1690032916 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 20 |
Hb_010863_030 |
0.1716383126 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |