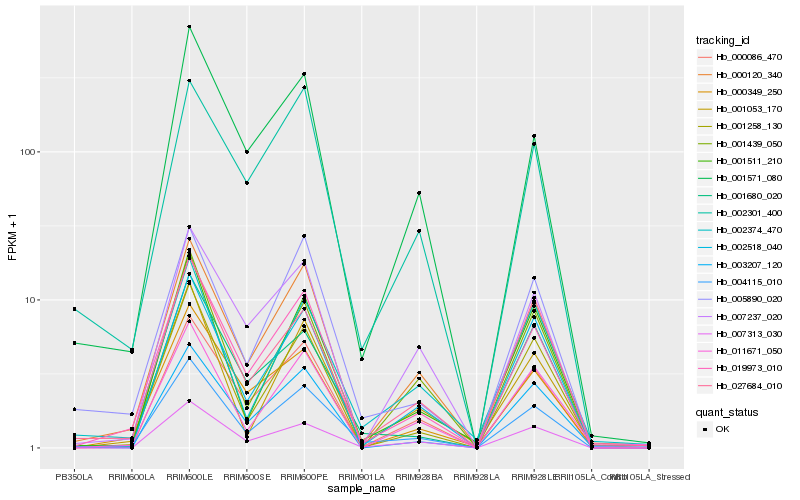
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004115_010 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 2 |
Hb_019973_010 |
0.0773937766 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000120_340 |
0.0959508673 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 4 |
Hb_007313_030 |
0.1005915052 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 5 |
Hb_000086_470 |
0.109715068 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 6 |
Hb_002518_040 |
0.1123144024 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 7 |
Hb_005890_020 |
0.1140656406 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 8 |
Hb_001571_080 |
0.1152157058 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 9 |
Hb_001258_130 |
0.1206928571 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 10 |
Hb_027684_010 |
0.123711668 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 11 |
Hb_001511_210 |
0.126471718 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 12 |
Hb_002374_470 |
0.1276943305 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_001680_020 |
0.1283339693 |
desease resistance |
Gene Name: RPW8 |
Disease resistance protein ADR1, putative [Ricinus communis] |
| 14 |
Hb_001053_170 |
0.1294930128 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 15 |
Hb_002301_400 |
0.1297143347 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 16 |
Hb_007237_020 |
0.1299065766 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 17 |
Hb_000349_250 |
0.1300124898 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 18 |
Hb_003207_120 |
0.1306650025 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 19 |
Hb_001439_050 |
0.1307370233 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 20 |
Hb_011671_050 |
0.1311202645 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |