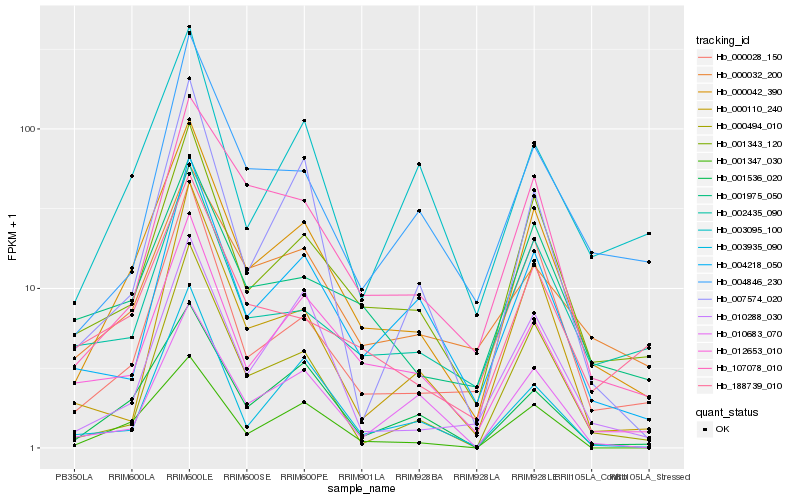
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_390 |
0.0 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7 [Jatropha curcas] |
| 2 |
Hb_001343_120 |
0.1027224209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001536_020 |
0.1280751427 |
transcription factor |
TF Family: GRF |
PREDICTED: uncharacterized protein LOC105631006 [Jatropha curcas] |
| 4 |
Hb_001347_030 |
0.1286490863 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_012653_010 |
0.1363071069 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 6 |
Hb_107078_010 |
0.148306746 |
- |
- |
- |
| 7 |
Hb_000494_010 |
0.1501946037 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0014s04460g [Populus trichocarpa] |
| 8 |
Hb_001975_050 |
0.1502263672 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |
| 9 |
Hb_004218_050 |
0.151162476 |
- |
- |
annexin [Manihot esculenta] |
| 10 |
Hb_000028_150 |
0.1572662632 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 11 |
Hb_010288_030 |
0.1586615748 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 12 |
Hb_004846_230 |
0.1631606858 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_188739_010 |
0.1634426364 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_000032_200 |
0.1644147965 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 15 |
Hb_007574_020 |
0.1659250009 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 16 |
Hb_003095_100 |
0.1665465176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002435_090 |
0.1667583467 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000110_240 |
0.1679135514 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 19 |
Hb_003935_090 |
0.1690912001 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 20 |
Hb_010683_070 |
0.1691831933 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT2 isoform X1 [Jatropha curcas] |