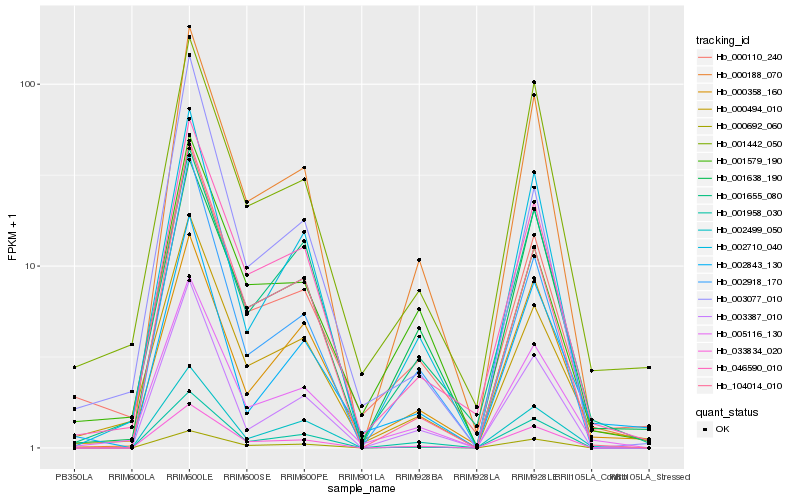
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005116_130 |
0.0 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000494_010 |
0.0759378049 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0014s04460g [Populus trichocarpa] |
| 3 |
Hb_046590_010 |
0.0905127215 |
- |
- |
Cycloeucalenol cycloisomerase, putative [Ricinus communis] |
| 4 |
Hb_000110_240 |
0.0924457348 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 5 |
Hb_000188_070 |
0.1039830364 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 6 |
Hb_001442_050 |
0.1096539755 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit U, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001655_080 |
0.110638788 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 8 |
Hb_002710_040 |
0.1135491623 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 9 |
Hb_033834_020 |
0.1152398155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001958_030 |
0.1201298719 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 11 |
Hb_002918_170 |
0.1208496889 |
- |
- |
PREDICTED: MACPF domain-containing protein NSL1 [Jatropha curcas] |
| 12 |
Hb_001638_190 |
0.1225691848 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000358_160 |
0.1228276111 |
- |
- |
PREDICTED: uncharacterized protein LOC105637769 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001579_190 |
0.1248345855 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000692_060 |
0.1271201417 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 16 |
Hb_003387_010 |
0.1276187555 |
- |
- |
PREDICTED: protein kinase PINOID 2 [Gossypium raimondii] |
| 17 |
Hb_002843_130 |
0.1306188175 |
- |
- |
PREDICTED: GRF1-interacting factor 2-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_002499_050 |
0.1309752949 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_104014_010 |
0.1338236129 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003077_010 |
0.1349635461 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |