| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
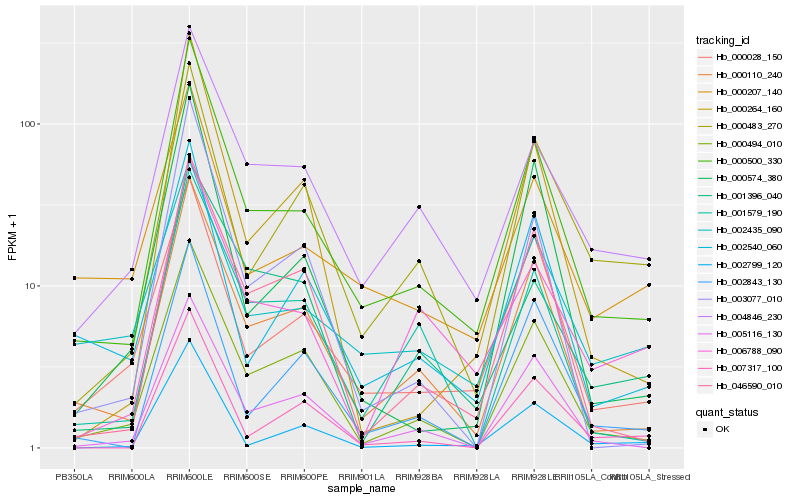
Hb_000500_330 |
0.0 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 2 |
Hb_000110_240 |
0.0973670754 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000494_010 |
0.1250680888 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0014s04460g [Populus trichocarpa] |
| 4 |
Hb_004846_230 |
0.1267255425 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_000028_150 |
0.1374526906 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 6 |
Hb_005116_130 |
0.1401346633 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 7 |
Hb_046590_010 |
0.1475527993 |
- |
- |
Cycloeucalenol cycloisomerase, putative [Ricinus communis] |
| 8 |
Hb_003077_010 |
0.1479043768 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000264_160 |
0.150914988 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 10 |
Hb_001579_190 |
0.1513073342 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002540_060 |
0.1514257023 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000207_140 |
0.1553537838 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 13 |
Hb_000574_380 |
0.1562073731 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 14 |
Hb_006788_090 |
0.1570109427 |
- |
- |
- |
| 15 |
Hb_000483_270 |
0.1576294107 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 16 |
Hb_002435_090 |
0.1625811437 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001396_040 |
0.1629617895 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 18 |
Hb_002799_120 |
0.1636859559 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 19 |
Hb_007317_100 |
0.1647824837 |
- |
- |
Peflin, putative [Ricinus communis] |
| 20 |
Hb_002843_130 |
0.1651281743 |
- |
- |
PREDICTED: GRF1-interacting factor 2-like isoform X2 [Jatropha curcas] |