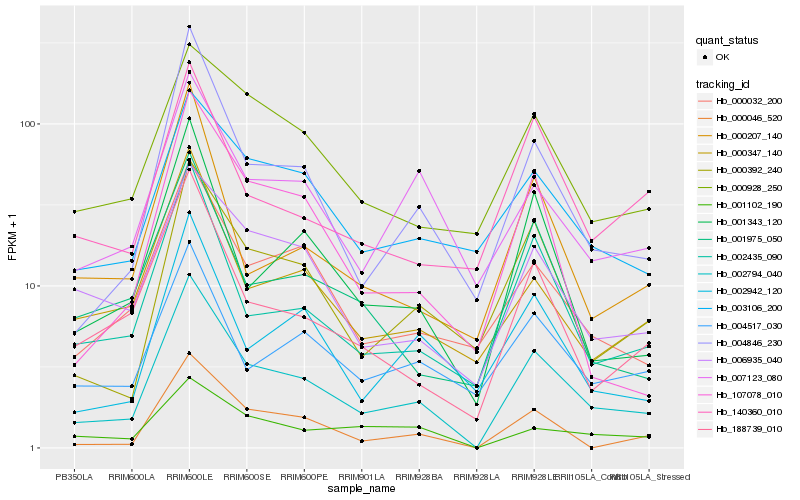
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002794_040 |
0.0 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 2 |
Hb_004846_230 |
0.1305920284 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_002435_090 |
0.1470036319 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000392_240 |
0.1496581168 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_188739_010 |
0.1521231753 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_001343_120 |
0.1555782484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000046_520 |
0.1582148516 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000032_200 |
0.1591952114 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 9 |
Hb_002942_120 |
0.1642884897 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 10 |
Hb_006935_040 |
0.1658976326 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 11 |
Hb_007123_080 |
0.1661992665 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 12 |
Hb_000347_140 |
0.1684465979 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_004517_030 |
0.171305273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_107078_010 |
0.1721284351 |
- |
- |
- |
| 15 |
Hb_001975_050 |
0.1731243199 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |
| 16 |
Hb_003106_200 |
0.1735507264 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001102_190 |
0.1750651914 |
- |
- |
PREDICTED: uncharacterized protein LOC105112724 [Populus euphratica] |
| 18 |
Hb_000207_140 |
0.1763408616 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 19 |
Hb_000928_250 |
0.1798205933 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_140360_010 |
0.1807619851 |
- |
- |
PREDICTED: 50S ribosomal protein L11, chloroplastic-like [Populus euphratica] |