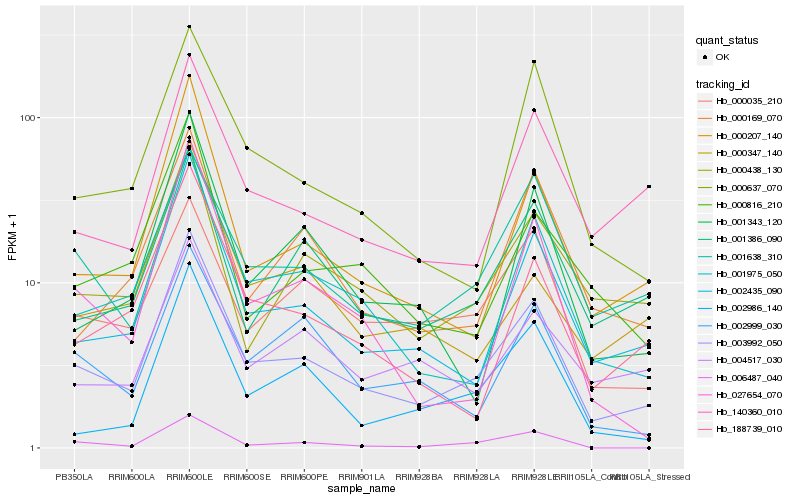
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003992_050 |
0.0 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 2 |
Hb_002435_090 |
0.1072991874 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000207_140 |
0.1209966663 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 4 |
Hb_001975_050 |
0.1222639153 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |
| 5 |
Hb_140360_010 |
0.1452743698 |
- |
- |
PREDICTED: 50S ribosomal protein L11, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_188739_010 |
0.1480503964 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_001386_090 |
0.1481102928 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 8 |
Hb_001638_310 |
0.1482313021 |
- |
- |
ribonucleoprotein, chloroplast, putative [Ricinus communis] |
| 9 |
Hb_000637_070 |
0.1490402413 |
- |
- |
PREDICTED: uncharacterized protein LOC105650668 [Jatropha curcas] |
| 10 |
Hb_027654_070 |
0.1520830779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001343_120 |
0.1568647806 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000816_210 |
0.1580663391 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase isoform X2 [Jatropha curcas] |
| 13 |
Hb_002999_030 |
0.1581363721 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 14 |
Hb_006487_040 |
0.1582013588 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 15 |
Hb_002986_140 |
0.1587746283 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 16 |
Hb_000169_070 |
0.1594940184 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 17 |
Hb_000035_210 |
0.1599395922 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 18 |
Hb_000438_130 |
0.159958916 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 19 |
Hb_004517_030 |
0.160724534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000347_140 |
0.1611340858 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |