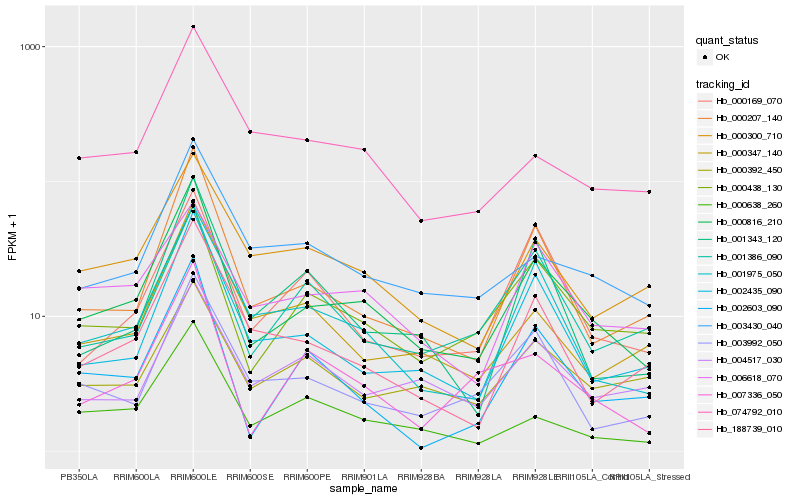
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_210 |
0.0 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase isoform X2 [Jatropha curcas] |
| 2 |
Hb_000207_140 |
0.1243226774 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 3 |
Hb_002435_090 |
0.1363897709 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001975_050 |
0.1367814122 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |
| 5 |
Hb_000438_130 |
0.1395205587 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 6 |
Hb_188739_010 |
0.1557926908 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_003430_040 |
0.1559096295 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 8 |
Hb_000300_710 |
0.1579762047 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 9 |
Hb_003992_050 |
0.1580663391 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 10 |
Hb_000638_260 |
0.1608234298 |
- |
- |
hypothetical protein JCGZ_06730 [Jatropha curcas] |
| 11 |
Hb_007336_050 |
0.1609598558 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 12 |
Hb_000347_140 |
0.1642736802 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_074792_010 |
0.1658579504 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 14 |
Hb_002603_090 |
0.1670350891 |
- |
- |
PREDICTED: GDSL esterase/lipase CPRD49-like isoform X2 [Populus euphratica] |
| 15 |
Hb_001343_120 |
0.1698678598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006618_070 |
0.1712477858 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000392_450 |
0.1715410956 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004517_030 |
0.1734146654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000169_070 |
0.1758969623 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 20 |
Hb_001386_090 |
0.1766642013 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |