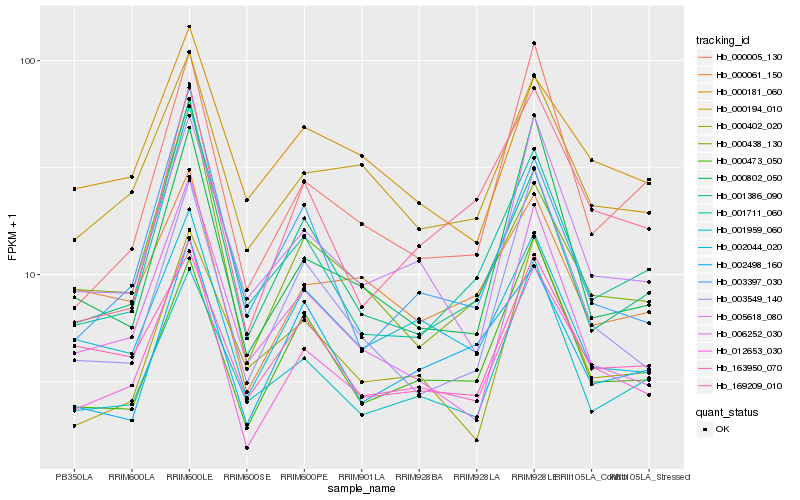
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012653_030 |
0.0 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 4, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_000194_010 |
0.1071102179 |
- |
- |
omega-6 fatty acid desaturase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005618_080 |
0.1081340825 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002044_020 |
0.1146156035 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 5 |
Hb_001711_060 |
0.1184791258 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 6 |
Hb_000438_130 |
0.1237918856 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 7 |
Hb_000473_050 |
0.1253211866 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 8 |
Hb_169209_010 |
0.1267285495 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_006252_030 |
0.1272203126 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 10 |
Hb_001386_090 |
0.128551134 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 11 |
Hb_001959_060 |
0.1288946236 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000005_130 |
0.1295981426 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 13 |
Hb_003549_140 |
0.130520986 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000802_050 |
0.1317770143 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 15 |
Hb_000181_060 |
0.1326444872 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000061_150 |
0.1329621463 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_163950_070 |
0.1349701226 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 18 |
Hb_003397_030 |
0.1351314599 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002498_160 |
0.1364600595 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 20 |
Hb_000402_020 |
0.1411332712 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |