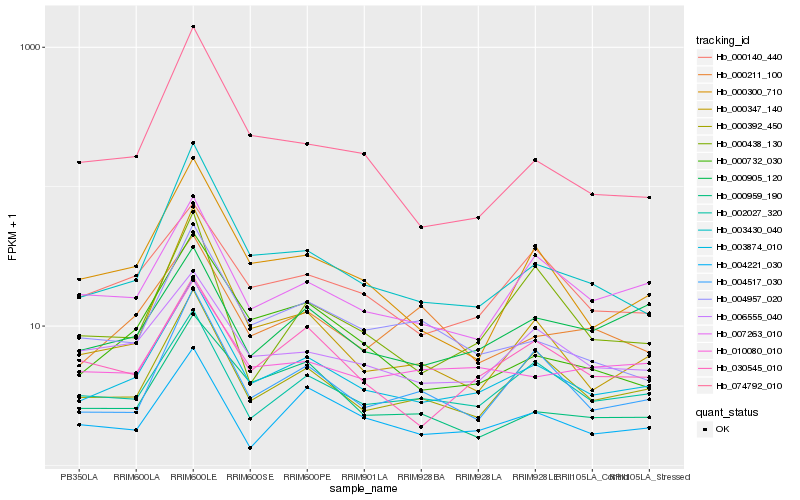
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003874_010 |
0.0 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_000392_450 |
0.0945044041 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_003430_040 |
0.1045007224 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 4 |
Hb_002027_320 |
0.113008155 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 5 |
Hb_007263_010 |
0.1142881598 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 6 |
Hb_004517_030 |
0.11613058 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000300_710 |
0.1205857051 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 8 |
Hb_006555_040 |
0.1208901724 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 9 |
Hb_030545_010 |
0.1229256721 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 10 |
Hb_010080_010 |
0.1279315447 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 11 |
Hb_000140_440 |
0.1291529098 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 12 |
Hb_004957_020 |
0.1292413928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004221_030 |
0.1315588055 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000905_120 |
0.1321746427 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 15 |
Hb_000732_030 |
0.1323252276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000347_140 |
0.1370285517 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_000959_190 |
0.1408356165 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 18 |
Hb_074792_010 |
0.1408852413 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 19 |
Hb_000211_100 |
0.1426745549 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 20 |
Hb_000438_130 |
0.1439696729 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |