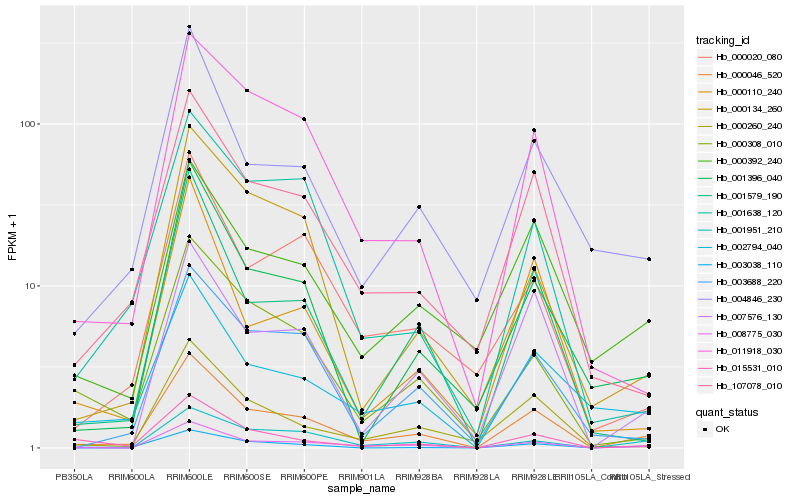
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_520 |
0.0 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_008775_030 |
0.0995945816 |
- |
- |
PREDICTED: ferric reduction oxidase 7, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_003038_110 |
0.1467984466 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 4 |
Hb_107078_010 |
0.1495056752 |
- |
- |
- |
| 5 |
Hb_002794_040 |
0.1582148516 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 6 |
Hb_007576_130 |
0.1614797964 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 7 |
Hb_000392_240 |
0.162529687 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_001396_040 |
0.1638831174 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 9 |
Hb_011918_030 |
0.1642645201 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 10 |
Hb_015531_010 |
0.1652167467 |
- |
- |
PREDICTED: uncharacterized protein LOC105636764 [Jatropha curcas] |
| 11 |
Hb_004846_230 |
0.1657731644 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_001579_190 |
0.1682551164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000308_010 |
0.1686198206 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 14 |
Hb_003688_220 |
0.1690004427 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_000260_240 |
0.1717788583 |
- |
- |
hypothetical protein JCGZ_00622 [Jatropha curcas] |
| 16 |
Hb_001951_210 |
0.1731136166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000020_080 |
0.177263496 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 18 |
Hb_001638_120 |
0.1804778356 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 19 |
Hb_000110_240 |
0.1806631181 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000134_260 |
0.1809290736 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |