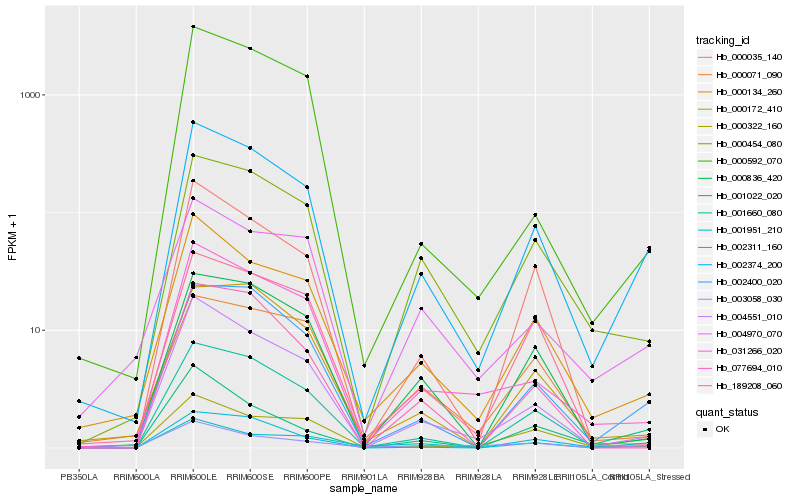
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_200 |
0.0 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 2 |
Hb_002400_020 |
0.1128116399 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 3 |
Hb_004551_010 |
0.1215099016 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 4 |
Hb_000134_260 |
0.1245491898 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000454_080 |
0.1312547066 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 6 |
Hb_001660_080 |
0.1366706757 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 7 |
Hb_000322_160 |
0.1403994706 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2-like [Jatropha curcas] |
| 8 |
Hb_002311_160 |
0.1410268345 |
transcription factor |
TF Family: ARR-B |
sensor histidine kinase, putative [Ricinus communis] |
| 9 |
Hb_031266_020 |
0.1464888356 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000836_420 |
0.1526468376 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 11 |
Hb_004970_070 |
0.1560868715 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 12 |
Hb_000172_410 |
0.1621393271 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 13 |
Hb_000035_140 |
0.1653820486 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 14 |
Hb_003058_030 |
0.1659270505 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 15 |
Hb_001022_020 |
0.1666412975 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 16 |
Hb_001951_210 |
0.1667479777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_189208_060 |
0.1670184534 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 18 |
Hb_077694_010 |
0.1733537498 |
- |
- |
uncharacterized protein LOC100305624 [Glycine max] |
| 19 |
Hb_000592_070 |
0.1737994125 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 20 |
Hb_000071_090 |
0.1741445506 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |