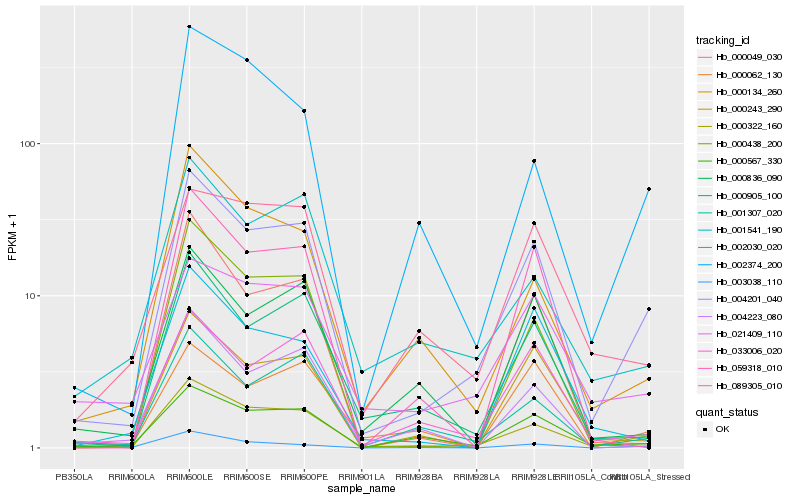
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004201_040 |
0.0 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 2 |
Hb_000322_160 |
0.0770882589 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2-like [Jatropha curcas] |
| 3 |
Hb_000567_330 |
0.1577676837 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 4 |
Hb_033006_020 |
0.1600713393 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |
| 5 |
Hb_001307_020 |
0.1642544232 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 6 |
Hb_000905_100 |
0.1688325441 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000243_290 |
0.1709066363 |
- |
- |
plastidic aldolase family protein [Populus trichocarpa] |
| 8 |
Hb_021409_110 |
0.1794621584 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_001541_190 |
0.1810687373 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 10 |
Hb_000134_260 |
0.1869166299 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000049_030 |
0.1873536418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000836_090 |
0.1881134128 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 13 |
Hb_002374_200 |
0.1886480983 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 14 |
Hb_003038_110 |
0.1905030035 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 15 |
Hb_089305_010 |
0.1920153127 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 16 |
Hb_000062_130 |
0.1923270724 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_059318_010 |
0.1932903173 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 18 |
Hb_000438_200 |
0.1964020676 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.1 [Jatropha curcas] |
| 19 |
Hb_004223_080 |
0.1968374214 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_002030_020 |
0.1979760909 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Jatropha curcas] |