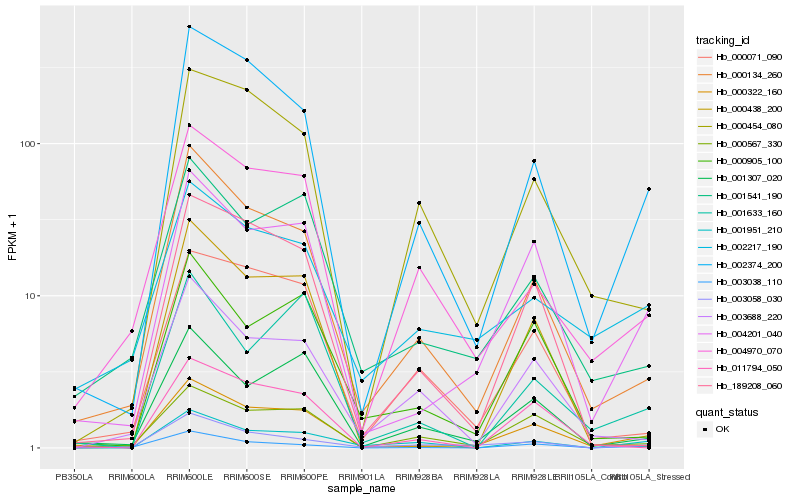
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000322_160 |
0.0 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2-like [Jatropha curcas] |
| 2 |
Hb_004201_040 |
0.0770882589 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 3 |
Hb_002374_200 |
0.1403994706 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 4 |
Hb_000134_260 |
0.1562890216 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_001541_190 |
0.1723458659 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 6 |
Hb_000567_330 |
0.1726950124 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 7 |
Hb_001307_020 |
0.1771936085 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 8 |
Hb_003058_030 |
0.1793747807 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000905_100 |
0.1843614619 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_189208_060 |
0.1863687987 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 11 |
Hb_000438_200 |
0.1879729845 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.1 [Jatropha curcas] |
| 12 |
Hb_001951_210 |
0.1886332849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004970_070 |
0.1887271206 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 14 |
Hb_000071_090 |
0.191729045 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 15 |
Hb_003038_110 |
0.1921944436 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 16 |
Hb_000454_080 |
0.1924910862 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 17 |
Hb_003688_220 |
0.1930987701 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_002217_190 |
0.1947236326 |
- |
- |
invertase [Hevea brasiliensis] |
| 19 |
Hb_011794_050 |
0.196018504 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_001633_160 |
0.1966553039 |
- |
- |
PREDICTED: zerumbone synthase-like isoform X2 [Jatropha curcas] |