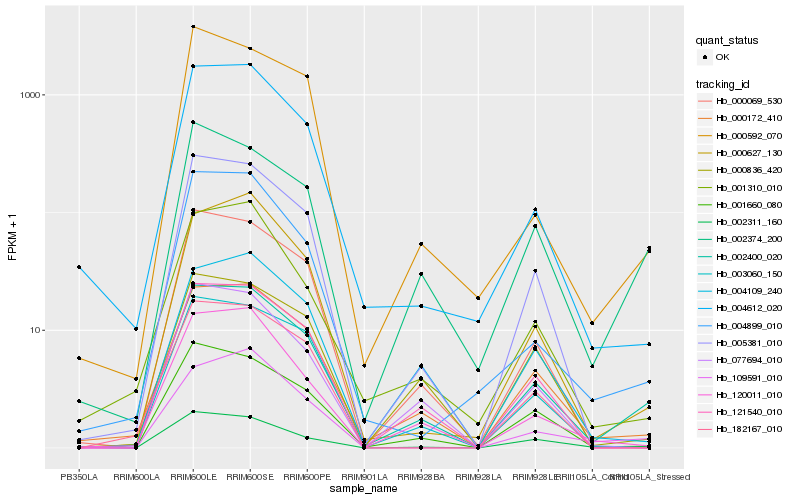
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_020 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 2 |
Hb_000172_410 |
0.0979012066 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_002374_200 |
0.1128116399 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 4 |
Hb_001660_080 |
0.1199341973 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 5 |
Hb_121540_010 |
0.1235292157 |
- |
- |
PREDICTED: UDP-glycosyltransferase 88A1-like [Jatropha curcas] |
| 6 |
Hb_002311_160 |
0.1238130772 |
transcription factor |
TF Family: ARR-B |
sensor histidine kinase, putative [Ricinus communis] |
| 7 |
Hb_005381_010 |
0.1241850348 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH35 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000069_530 |
0.1311925117 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0014s09190g [Populus trichocarpa] |
| 9 |
Hb_003060_150 |
0.1338389622 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_120011_010 |
0.1374259675 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_077694_010 |
0.1391084141 |
- |
- |
uncharacterized protein LOC100305624 [Glycine max] |
| 12 |
Hb_000836_420 |
0.1400540196 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000627_130 |
0.1407312703 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 14 |
Hb_109591_010 |
0.1414317255 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 15 |
Hb_004612_020 |
0.1416151996 |
- |
- |
PREDICTED: tetraspanin-8-like [Populus euphratica] |
| 16 |
Hb_004109_240 |
0.1441778635 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 17 |
Hb_000592_070 |
0.1463402708 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 18 |
Hb_001310_010 |
0.1475541887 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type [Jatropha curcas] |
| 19 |
Hb_004899_010 |
0.1486755876 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 20 |
Hb_182167_010 |
0.1511000071 |
- |
- |
hypothetical protein PHAVU_001G0486001g, partial [Phaseolus vulgaris] |