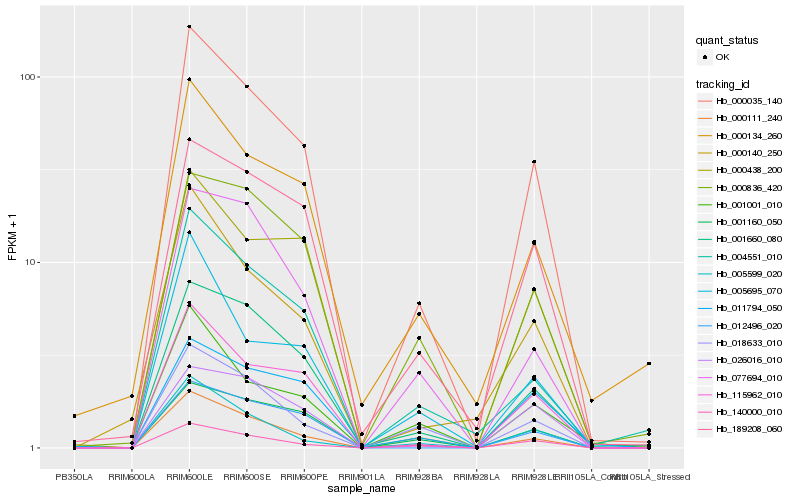
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_140 |
0.0 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 2 |
Hb_115962_010 |
0.0947075849 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 3 |
Hb_001660_080 |
0.0962470545 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 4 |
Hb_189208_060 |
0.1105708876 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 5 |
Hb_001160_050 |
0.1210638422 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 6 |
Hb_004551_010 |
0.1219061013 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 7 |
Hb_012496_020 |
0.1251254017 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 8 |
Hb_000140_250 |
0.1260895283 |
- |
- |
Late embryogenesis abundant protein Lea14-A, putative [Ricinus communis] |
| 9 |
Hb_000438_200 |
0.1299134328 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.1 [Jatropha curcas] |
| 10 |
Hb_026016_010 |
0.1307912849 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_140000_010 |
0.1326843459 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 12 |
Hb_011794_050 |
0.13283029 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_018633_010 |
0.1330412643 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000134_260 |
0.1341189447 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_001001_010 |
0.1355537159 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 16 |
Hb_005599_020 |
0.1374243132 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 17 |
Hb_000836_420 |
0.1375150643 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_000111_240 |
0.1415997227 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Elaeis guineensis] |
| 19 |
Hb_077694_010 |
0.1416212342 |
- |
- |
uncharacterized protein LOC100305624 [Glycine max] |
| 20 |
Hb_005695_070 |
0.1441504208 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |