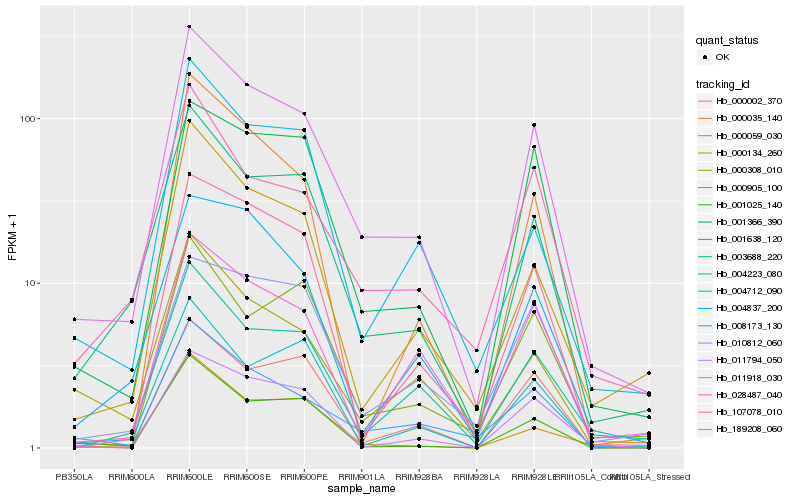
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011918_030 |
0.0 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 2 |
Hb_001638_120 |
0.1011775987 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 3 |
Hb_008173_130 |
0.1052107245 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003688_220 |
0.1237918645 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_107078_010 |
0.1246320746 |
- |
- |
- |
| 6 |
Hb_000134_260 |
0.1282962652 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_001366_390 |
0.1358863513 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 8 |
Hb_189208_060 |
0.1368950873 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 9 |
Hb_000308_010 |
0.1381263989 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 10 |
Hb_011794_050 |
0.1400528422 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_004712_090 |
0.1413544182 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 12 |
Hb_001025_140 |
0.1430603606 |
- |
- |
hypothetical protein POPTR_0015s09180g [Populus trichocarpa] |
| 13 |
Hb_028487_040 |
0.1460572091 |
- |
- |
hypothetical protein POPTR_0010s11910g [Populus trichocarpa] |
| 14 |
Hb_000002_370 |
0.1471485032 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 15 |
Hb_000905_100 |
0.1484120905 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_004837_200 |
0.1485161005 |
- |
- |
hypothetical protein POPTR_0001s26450g [Populus trichocarpa] |
| 17 |
Hb_000035_140 |
0.1535541283 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 18 |
Hb_000059_030 |
0.1538023377 |
- |
- |
PREDICTED: chaperone protein dnaJ 10 [Jatropha curcas] |
| 19 |
Hb_010812_060 |
0.1547588925 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 20 |
Hb_004223_080 |
0.1567931896 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |