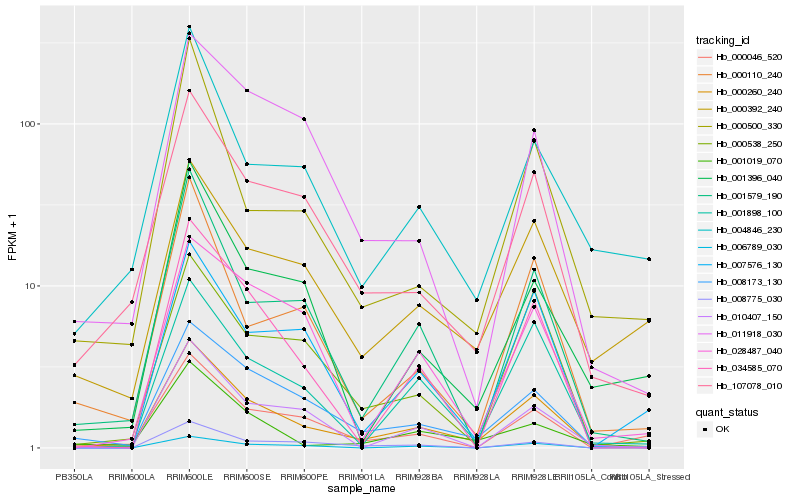
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_240 |
0.0 |
- |
- |
hypothetical protein JCGZ_00622 [Jatropha curcas] |
| 2 |
Hb_008173_130 |
0.1209443918 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 3 |
Hb_107078_010 |
0.1331977571 |
- |
- |
- |
| 4 |
Hb_001898_100 |
0.1434520903 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 5 |
Hb_001579_190 |
0.1483592063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_034585_070 |
0.1486421054 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 7 |
Hb_001019_070 |
0.1619782527 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 8 |
Hb_000538_250 |
0.162979759 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 [Jatropha curcas] |
| 9 |
Hb_000110_240 |
0.1633441438 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000392_240 |
0.1705692511 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_004846_230 |
0.17134482 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000046_520 |
0.1717788583 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001396_040 |
0.1748371541 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 14 |
Hb_006789_030 |
0.1751492378 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Prunus mume] |
| 15 |
Hb_000500_330 |
0.1758176749 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 16 |
Hb_011918_030 |
0.1784840606 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 17 |
Hb_007576_130 |
0.1795986338 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 18 |
Hb_008775_030 |
0.1806615059 |
- |
- |
PREDICTED: ferric reduction oxidase 7, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_010407_150 |
0.1807617713 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 20 |
Hb_028487_040 |
0.1820870868 |
- |
- |
hypothetical protein POPTR_0010s11910g [Populus trichocarpa] |