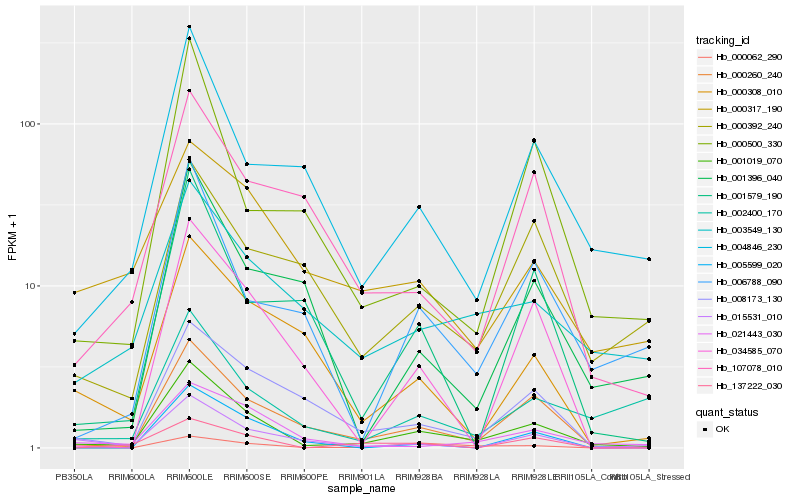
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001019_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 2 |
Hb_000260_240 |
0.1619782527 |
- |
- |
hypothetical protein JCGZ_00622 [Jatropha curcas] |
| 3 |
Hb_004846_230 |
0.211288316 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_003549_130 |
0.2149730791 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 5 |
Hb_021443_030 |
0.217216789 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 6 |
Hb_006788_090 |
0.219952847 |
- |
- |
- |
| 7 |
Hb_008173_130 |
0.2217136475 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002400_170 |
0.2276355813 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 9 |
Hb_000500_330 |
0.2351196402 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 10 |
Hb_015531_010 |
0.2351707202 |
- |
- |
PREDICTED: uncharacterized protein LOC105636764 [Jatropha curcas] |
| 11 |
Hb_034585_070 |
0.2352616791 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 12 |
Hb_001396_040 |
0.2366268864 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 13 |
Hb_000062_290 |
0.2366868156 |
desease resistance |
Gene Name: ABC_membrane |
hypothetical protein POPTR_0008s17960g [Populus trichocarpa] |
| 14 |
Hb_107078_010 |
0.2407632302 |
- |
- |
- |
| 15 |
Hb_000308_010 |
0.2415968474 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 16 |
Hb_001579_190 |
0.2423790178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_137222_030 |
0.243389126 |
- |
- |
- |
| 18 |
Hb_000392_240 |
0.2446051842 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_005599_020 |
0.2456853231 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 20 |
Hb_000317_190 |
0.2469710608 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |