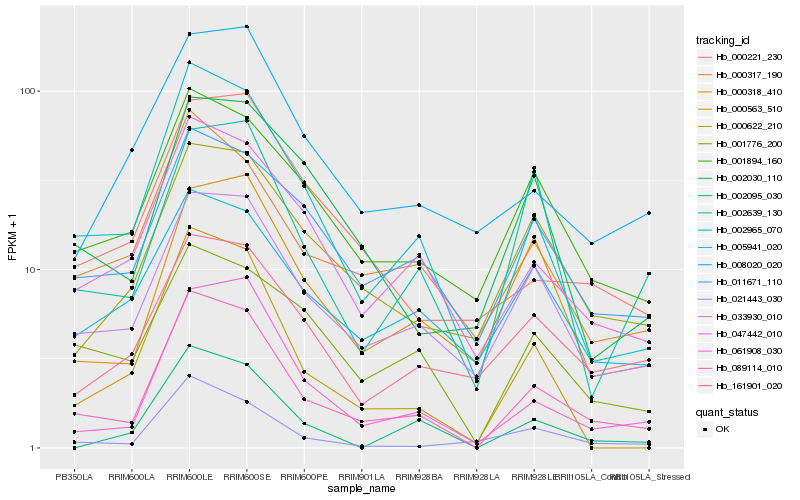
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089114_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 2 |
Hb_002965_070 |
0.122780503 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 3 |
Hb_061908_030 |
0.142637984 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 4 |
Hb_033930_010 |
0.1479655721 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 5 |
Hb_047442_010 |
0.1491114151 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 6 |
Hb_000317_190 |
0.1583457355 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 7 |
Hb_001894_160 |
0.1632987803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000563_510 |
0.1708033817 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 9 |
Hb_011671_110 |
0.1726870912 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 10 |
Hb_000622_210 |
0.1740646649 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 11 |
Hb_001776_200 |
0.1753711401 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 12 |
Hb_021443_030 |
0.1755201073 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 13 |
Hb_000318_410 |
0.1784089794 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 14 |
Hb_005941_020 |
0.184718223 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 15 |
Hb_000221_230 |
0.1869936347 |
transcription factor |
TF Family: Jumonji |
PREDICTED: F-box protein At1g78280 [Jatropha curcas] |
| 16 |
Hb_002030_110 |
0.1874252347 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 17 |
Hb_161901_020 |
0.188187838 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 18 |
Hb_008020_020 |
0.1889285171 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002639_130 |
0.1918565904 |
- |
- |
hypothetical protein JCGZ_08129 [Jatropha curcas] |
| 20 |
Hb_002095_030 |
0.1922744747 |
- |
- |
hypothetical protein MIMGU_mgv1a021906mg, partial [Erythranthe guttata] |