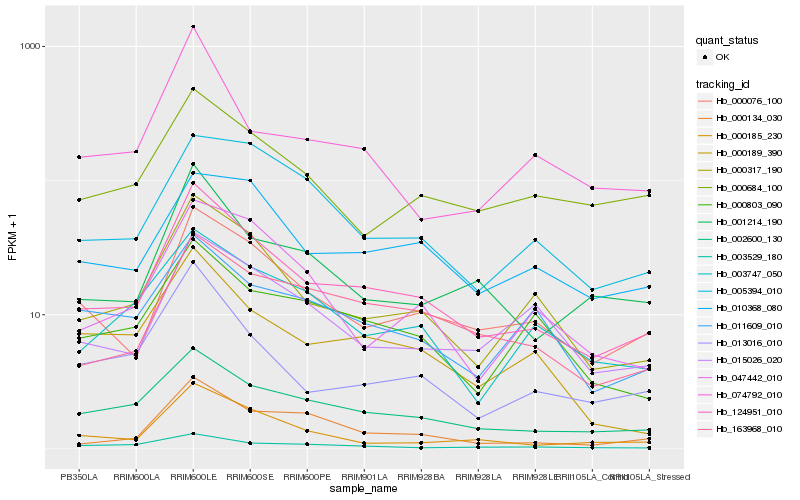
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_124951_010 |
0.0 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 2 |
Hb_002600_130 |
0.1071727138 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 3 |
Hb_000317_190 |
0.1078667339 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 4 |
Hb_000076_100 |
0.111528184 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1 [Jatropha curcas] |
| 5 |
Hb_163968_010 |
0.1369499522 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 6 |
Hb_001214_190 |
0.1382417553 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 7 |
Hb_000134_030 |
0.1386182101 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 8 |
Hb_013016_010 |
0.1399980713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000185_230 |
0.1434393707 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 10 |
Hb_003529_180 |
0.1454361706 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 11 |
Hb_003747_050 |
0.1465386995 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 12 |
Hb_000189_390 |
0.1475667734 |
- |
- |
PREDICTED: pollen-specific protein SF21-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_015026_020 |
0.1496598221 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 14 |
Hb_010368_080 |
0.1524626778 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 15 |
Hb_000684_100 |
0.1528627253 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 16 |
Hb_000803_090 |
0.1548177348 |
- |
- |
PREDICTED: transcription initiation factor IIB isoform X1 [Jatropha curcas] |
| 17 |
Hb_047442_010 |
0.1575006322 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 18 |
Hb_011609_010 |
0.15973485 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 19 |
Hb_005394_010 |
0.1601415834 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 20 |
Hb_074792_010 |
0.1618327074 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |