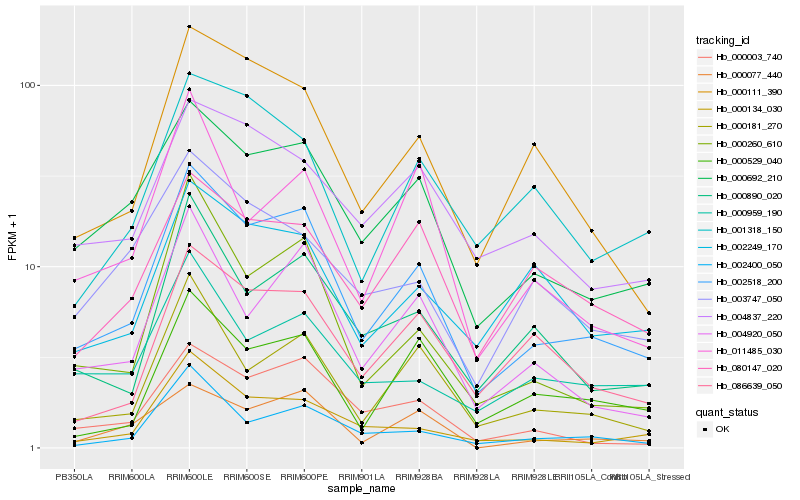
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002518_200 |
0.0 |
- |
- |
Proliferating cell nuclear antigen [Theobroma cacao] |
| 2 |
Hb_000692_210 |
0.1131455324 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2 [Jatropha curcas] |
| 3 |
Hb_000181_270 |
0.130175819 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_004920_050 |
0.1322156435 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 5 |
Hb_000003_740 |
0.1328668905 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 6 |
Hb_000111_390 |
0.1374148938 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000134_030 |
0.1401492172 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 8 |
Hb_000890_020 |
0.1408492361 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 9 |
Hb_000260_610 |
0.1424328743 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 10 |
Hb_000077_440 |
0.1442744641 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 11 |
Hb_002400_050 |
0.1447046461 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 12 |
Hb_086639_050 |
0.1486785849 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000529_040 |
0.1496915004 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |
| 14 |
Hb_004837_220 |
0.1576933387 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 15 |
Hb_080147_020 |
0.1586760539 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 16 |
Hb_000959_190 |
0.1597768704 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 17 |
Hb_002249_170 |
0.1625895766 |
- |
- |
copine, putative [Ricinus communis] |
| 18 |
Hb_011485_030 |
0.1656245056 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 19 |
Hb_001318_150 |
0.1657210815 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 20 |
Hb_003747_050 |
0.1678292901 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |