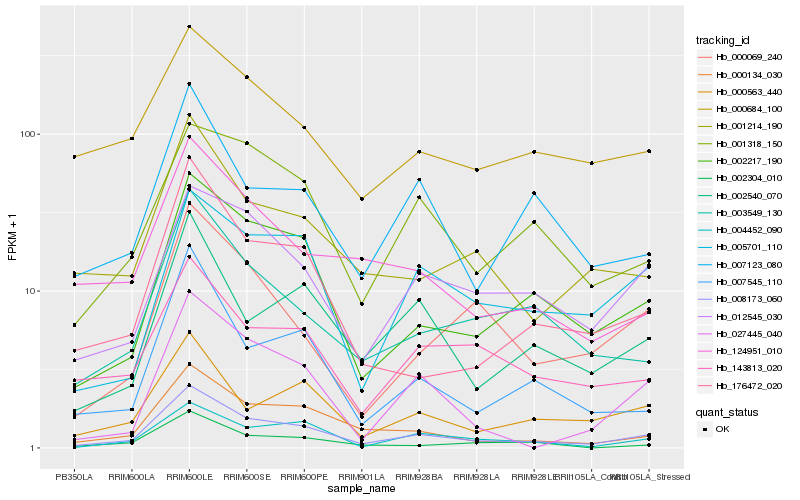
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008173_060 |
0.0 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 2 |
Hb_002217_190 |
0.1239786554 |
- |
- |
invertase [Hevea brasiliensis] |
| 3 |
Hb_000134_030 |
0.1288140082 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 4 |
Hb_004452_090 |
0.1425869018 |
- |
- |
hypothetical protein JCGZ_12231 [Jatropha curcas] |
| 5 |
Hb_176472_020 |
0.1505416364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007545_110 |
0.1566662597 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_012545_030 |
0.1566721273 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 8 |
Hb_003549_130 |
0.1579798873 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 9 |
Hb_002540_070 |
0.15909068 |
- |
- |
PREDICTED: protein cornichon homolog 1 [Jatropha curcas] |
| 10 |
Hb_001214_190 |
0.159871514 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 11 |
Hb_000563_440 |
0.1608518692 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001318_150 |
0.16454255 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 13 |
Hb_027445_040 |
0.1646314581 |
- |
- |
PREDICTED: uncharacterized protein LOC105117996 [Populus euphratica] |
| 14 |
Hb_143813_020 |
0.1664419747 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 15 |
Hb_005701_110 |
0.1691790491 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 16 |
Hb_124951_010 |
0.1698699988 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 17 |
Hb_000069_240 |
0.1725272239 |
- |
- |
PREDICTED: protein DEFECTIVE IN MERISTEM SILENCING 3-like [Jatropha curcas] |
| 18 |
Hb_007123_080 |
0.1731738035 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 19 |
Hb_000684_100 |
0.1749520129 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 20 |
Hb_002304_010 |
0.1752505918 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |