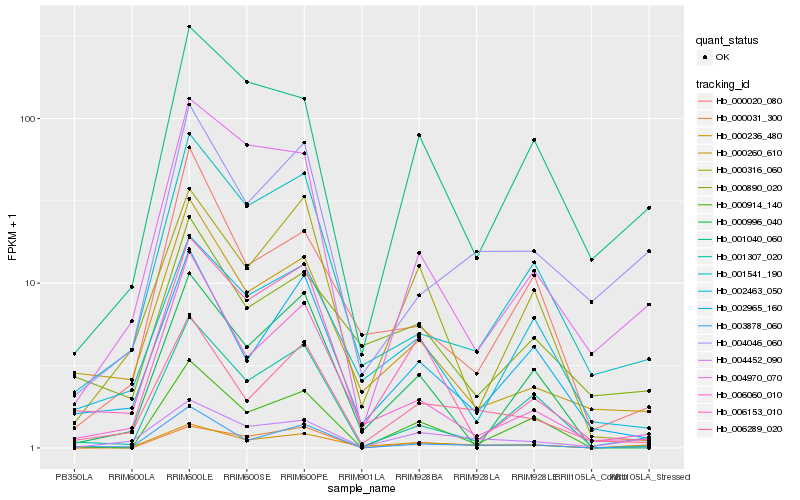
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000236_480 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Jatropha curcas] |
| 2 |
Hb_006289_020 |
0.155345421 |
- |
- |
PREDICTED: shugoshin-1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004452_090 |
0.1598120065 |
- |
- |
hypothetical protein JCGZ_12231 [Jatropha curcas] |
| 4 |
Hb_000031_300 |
0.1724895883 |
- |
- |
PREDICTED: histidine kinase 5 [Jatropha curcas] |
| 5 |
Hb_001541_190 |
0.1758985327 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 6 |
Hb_006060_010 |
0.1791066618 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 7 |
Hb_000020_080 |
0.1805921294 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 8 |
Hb_002463_050 |
0.1867158355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004046_060 |
0.1868400494 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 10 |
Hb_000890_020 |
0.1897364789 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 11 |
Hb_001040_060 |
0.1903919277 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 12 |
Hb_000260_610 |
0.1908755664 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 13 |
Hb_002965_160 |
0.192366221 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 14 |
Hb_000914_140 |
0.1929374531 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_006153_010 |
0.1930391376 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 [Populus euphratica] |
| 16 |
Hb_000316_060 |
0.1935038885 |
- |
- |
hypothetical protein CISIN_1g017081mg [Citrus sinensis] |
| 17 |
Hb_004970_070 |
0.194024074 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 18 |
Hb_003878_060 |
0.1948776638 |
- |
- |
hypothetical protein B456_007G326900, partial [Gossypium raimondii] |
| 19 |
Hb_001307_020 |
0.1963959502 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 20 |
Hb_000996_040 |
0.2004953007 |
- |
- |
hypothetical protein POPTR_0003s12500g [Populus trichocarpa] |