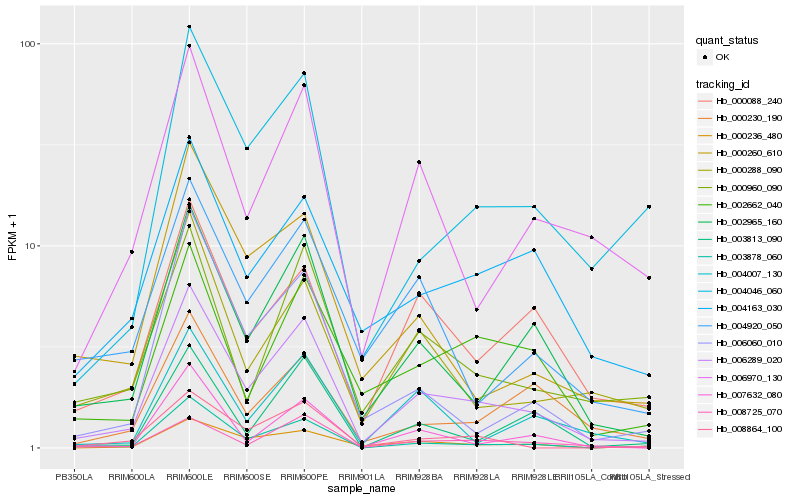
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006289_020 |
0.0 |
- |
- |
PREDICTED: shugoshin-1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002965_160 |
0.1283253458 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 3 |
Hb_003878_060 |
0.1451560157 |
- |
- |
hypothetical protein B456_007G326900, partial [Gossypium raimondii] |
| 4 |
Hb_000236_480 |
0.155345421 |
- |
- |
PREDICTED: cytochrome P450 CYP72A219-like [Jatropha curcas] |
| 5 |
Hb_008725_070 |
0.162288441 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 6 |
Hb_000960_090 |
0.1664213786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000288_090 |
0.1682296223 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000088_240 |
0.1685559136 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_006970_130 |
0.1698108784 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 10 |
Hb_004046_060 |
0.171286096 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 11 |
Hb_000260_610 |
0.1745724198 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 12 |
Hb_006060_010 |
0.1745874435 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 13 |
Hb_008864_100 |
0.175559012 |
- |
- |
PREDICTED: uncharacterized protein LOC105631832 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004007_130 |
0.1779554205 |
- |
- |
PREDICTED: uncharacterized protein LOC105639843 [Jatropha curcas] |
| 15 |
Hb_002662_040 |
0.1780307981 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 16 |
Hb_000230_190 |
0.1787867847 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004163_030 |
0.1788597516 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 18 |
Hb_007632_080 |
0.1813795691 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 19 |
Hb_003813_090 |
0.1830302783 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 20 |
Hb_004920_050 |
0.1833320851 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |