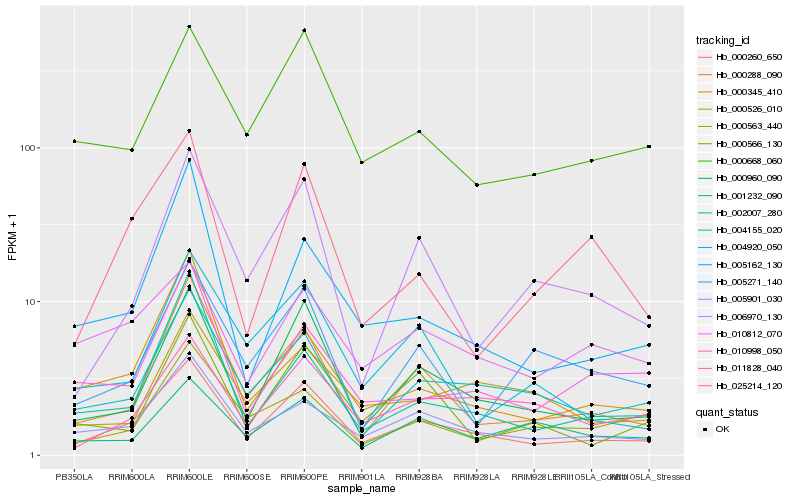
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_650 |
0.0 |
- |
- |
PREDICTED: protein PFC0760c [Jatropha curcas] |
| 2 |
Hb_000960_090 |
0.1114322158 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005901_030 |
0.1286517852 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004155_020 |
0.1311220476 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 5 |
Hb_000288_090 |
0.1355730438 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000668_060 |
0.1511768388 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 7 |
Hb_006970_130 |
0.1628197236 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 8 |
Hb_000345_410 |
0.1639019177 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 9 |
Hb_025214_120 |
0.1660810713 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |
| 10 |
Hb_011828_040 |
0.1677313928 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000566_130 |
0.1728656052 |
- |
- |
PREDICTED: probable DNA primase large subunit isoform X2 [Jatropha curcas] |
| 12 |
Hb_004920_050 |
0.1741622138 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 13 |
Hb_010812_070 |
0.1746116651 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein NETWORKED 4A [Jatropha curcas] |
| 14 |
Hb_005162_130 |
0.1746969083 |
- |
- |
PREDICTED: uncharacterized protein LOC105648433 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002007_280 |
0.1751132924 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 16 |
Hb_005271_140 |
0.1798464144 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3 [Morus notabilis] |
| 17 |
Hb_010998_050 |
0.1815235881 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000563_440 |
0.1826533906 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001232_090 |
0.1843546813 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 20 |
Hb_000526_010 |
0.1847637991 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |