| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
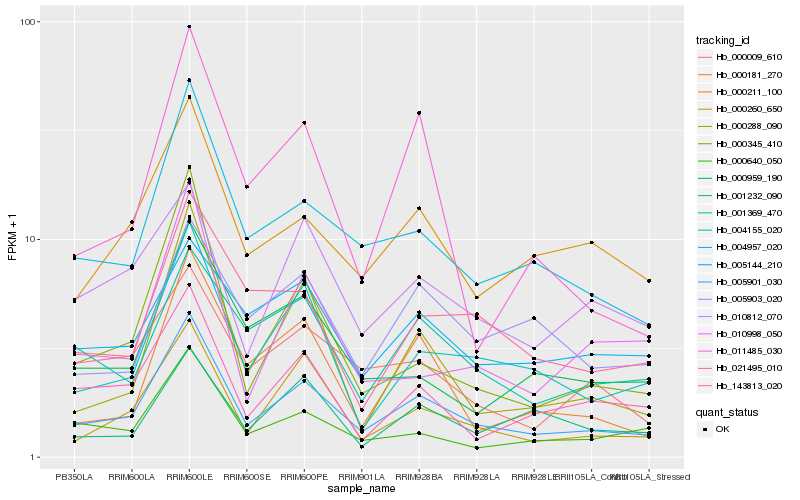
Hb_005901_030 |
0.0 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004957_020 |
0.1075855959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004155_020 |
0.1226845698 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 4 |
Hb_000260_650 |
0.1286517852 |
- |
- |
PREDICTED: protein PFC0760c [Jatropha curcas] |
| 5 |
Hb_000211_100 |
0.1310639347 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 6 |
Hb_021495_010 |
0.1412135042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000640_050 |
0.1439622875 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 8 |
Hb_001369_470 |
0.147618192 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 9 |
Hb_000345_410 |
0.1477074887 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 10 |
Hb_000288_090 |
0.1495600243 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_143813_020 |
0.153539686 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 12 |
Hb_000959_190 |
0.1541585138 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 13 |
Hb_010812_070 |
0.1545772435 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein NETWORKED 4A [Jatropha curcas] |
| 14 |
Hb_000181_270 |
0.1598181074 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_005903_020 |
0.1603590165 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 16 |
Hb_000009_610 |
0.1606391358 |
- |
- |
PREDICTED: uncharacterized protein LOC105639638 isoform X2 [Jatropha curcas] |
| 17 |
Hb_011485_030 |
0.1612425007 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 18 |
Hb_005144_210 |
0.1622769284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010998_050 |
0.1629485361 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001232_090 |
0.1636739032 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |