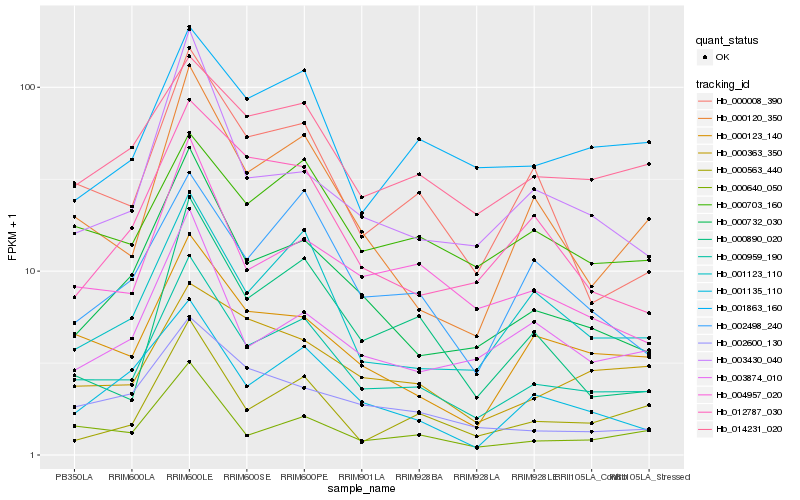
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000959_190 |
0.0 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 2 |
Hb_000732_030 |
0.1089973228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001123_110 |
0.1141449069 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000120_350 |
0.1164640786 |
- |
- |
PREDICTED: uncharacterized protein LOC104422872 isoform X1 [Eucalyptus grandis] |
| 5 |
Hb_000008_390 |
0.1187663927 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 6 |
Hb_000640_050 |
0.1191647324 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 7 |
Hb_004957_020 |
0.1197505214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000123_140 |
0.1234706845 |
- |
- |
PREDICTED: 21 kDa protein-like [Populus euphratica] |
| 9 |
Hb_014231_020 |
0.1285561331 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 10 |
Hb_001135_110 |
0.1338928547 |
- |
- |
- |
| 11 |
Hb_000703_160 |
0.1352814187 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001863_160 |
0.1378716268 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 13 |
Hb_002600_130 |
0.1390969641 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 14 |
Hb_003874_010 |
0.1408356165 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_002498_240 |
0.1408999292 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000563_440 |
0.1423865802 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003430_040 |
0.1426210147 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 18 |
Hb_012787_030 |
0.1439089547 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 19 |
Hb_000890_020 |
0.1461729947 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 20 |
Hb_000363_350 |
0.1476393964 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like [Fragaria vesca subsp. vesca] |