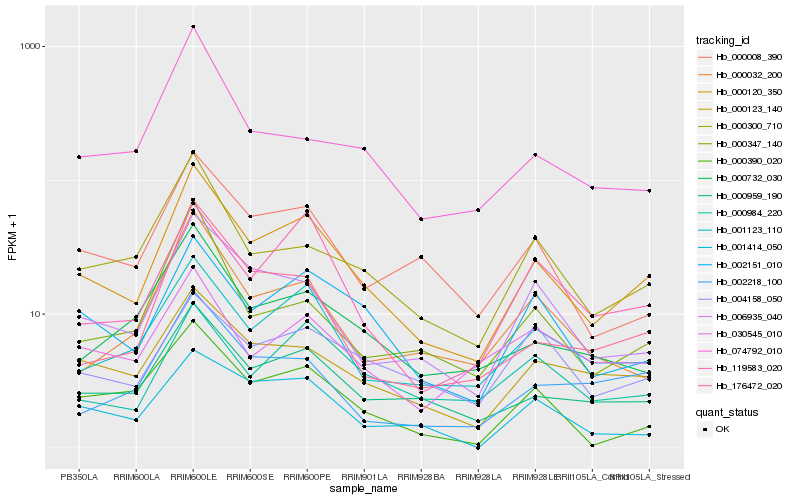
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_350 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104422872 isoform X1 [Eucalyptus grandis] |
| 2 |
Hb_000959_190 |
0.1164640786 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 3 |
Hb_001123_110 |
0.1290760914 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002151_010 |
0.1306284079 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 5 |
Hb_000123_140 |
0.1318865372 |
- |
- |
PREDICTED: 21 kDa protein-like [Populus euphratica] |
| 6 |
Hb_006935_040 |
0.133507956 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 7 |
Hb_000300_710 |
0.139145975 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 8 |
Hb_000008_390 |
0.1475757243 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 9 |
Hb_000390_020 |
0.1534159403 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 4-like [Jatropha curcas] |
| 10 |
Hb_000732_030 |
0.1540043676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_119583_020 |
0.1547446738 |
- |
- |
PREDICTED: uncharacterized protein LOC105628090 [Jatropha curcas] |
| 12 |
Hb_176472_020 |
0.1602867677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000984_220 |
0.1617359015 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 14 |
Hb_001414_050 |
0.1626672727 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 15 |
Hb_002218_100 |
0.1628628557 |
- |
- |
PREDICTED: uncharacterized protein At2g34160 [Jatropha curcas] |
| 16 |
Hb_000032_200 |
0.1632690235 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 17 |
Hb_030545_010 |
0.1657865737 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 18 |
Hb_000347_140 |
0.1658296076 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_074792_010 |
0.167363573 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 20 |
Hb_004158_050 |
0.1679586669 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |