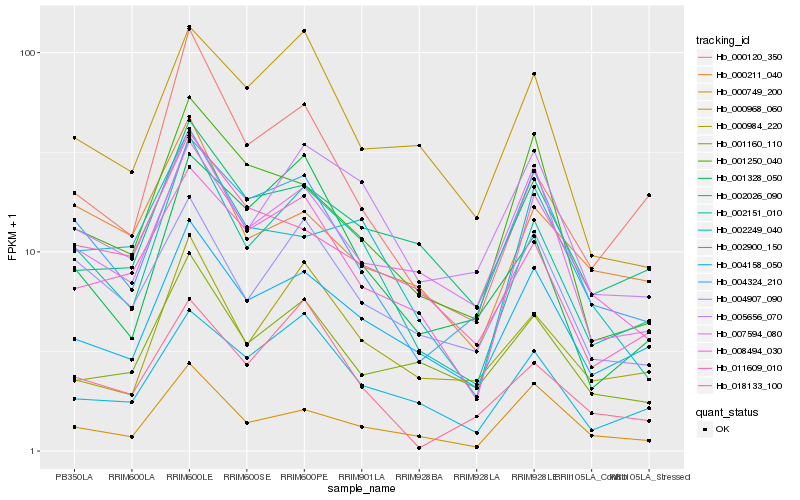
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002151_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 2 |
Hb_004158_050 |
0.1210158435 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 3 |
Hb_000120_350 |
0.1306284079 |
- |
- |
PREDICTED: uncharacterized protein LOC104422872 isoform X1 [Eucalyptus grandis] |
| 4 |
Hb_002900_150 |
0.1352249961 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 5 |
Hb_004324_210 |
0.1383742153 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 6 |
Hb_004907_090 |
0.1383964003 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_000984_220 |
0.142161645 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 8 |
Hb_005656_070 |
0.1483860841 |
- |
- |
PREDICTED: cryptochrome DASH, chloroplastic/mitochondrial [Jatropha curcas] |
| 9 |
Hb_018133_100 |
0.1493494673 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_001328_050 |
0.1519307927 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 11 |
Hb_000749_200 |
0.1548725772 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007594_080 |
0.1553668686 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 13 |
Hb_002026_090 |
0.1566697834 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 14 |
Hb_008494_030 |
0.1601179977 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_000211_040 |
0.1613562738 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 16 |
Hb_011609_010 |
0.1616085318 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 17 |
Hb_002249_040 |
0.1629487983 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1-like [Jatropha curcas] |
| 18 |
Hb_000968_060 |
0.1631445008 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 19 |
Hb_001160_110 |
0.1636529123 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 20 |
Hb_001250_040 |
0.1641968113 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |