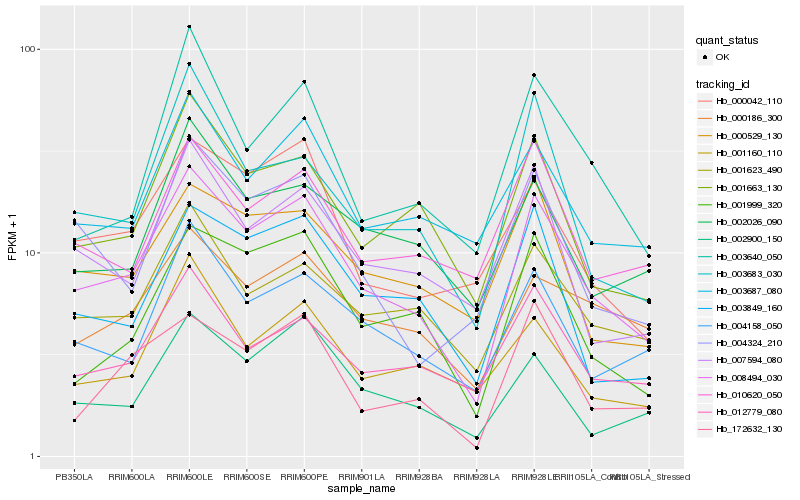
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008494_030 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_001663_130 |
0.1159934393 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 3 |
Hb_004158_050 |
0.1177772946 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 4 |
Hb_001623_490 |
0.1192294708 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001999_320 |
0.1218666022 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 6 |
Hb_003849_160 |
0.1224313106 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 7 |
Hb_003687_080 |
0.1236999104 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 8 |
Hb_012779_080 |
0.1249127387 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 9 |
Hb_002900_150 |
0.1251168322 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |
| 10 |
Hb_007594_080 |
0.1257845939 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 11 |
Hb_000529_130 |
0.1275046735 |
- |
- |
hypothetical protein POPTR_0007s14320g [Populus trichocarpa] |
| 12 |
Hb_000042_110 |
0.1282723314 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 13 |
Hb_000186_300 |
0.128859431 |
- |
- |
PREDICTED: malonyl-CoA decarboxylase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_172632_130 |
0.1294528724 |
- |
- |
hypothetical protein JCGZ_22584 [Jatropha curcas] |
| 15 |
Hb_003683_030 |
0.1307496822 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 16 |
Hb_002026_090 |
0.1312564119 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 17 |
Hb_010620_050 |
0.1312672743 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 18 |
Hb_004324_210 |
0.1322166179 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 19 |
Hb_003640_050 |
0.1351574938 |
- |
- |
PREDICTED: protein LUTEIN DEFICIENT 5, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_001160_110 |
0.1352091251 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |