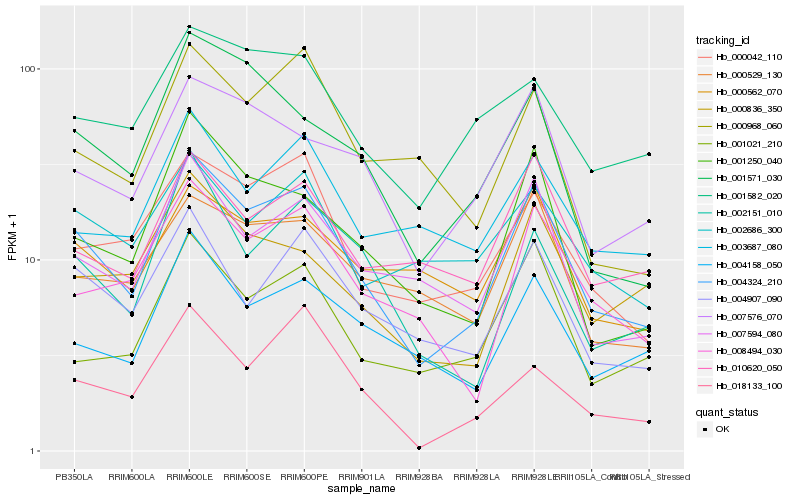
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004324_210 |
0.0 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 2 |
Hb_007594_080 |
0.1203175224 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 3 |
Hb_001250_040 |
0.1213034977 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 4 |
Hb_004907_090 |
0.1234754912 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000042_110 |
0.1300989058 |
- |
- |
PREDICTED: receptor-like protein kinase THESEUS 1 [Jatropha curcas] |
| 6 |
Hb_007576_070 |
0.1317737817 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X3 [Jatropha curcas] |
| 7 |
Hb_008494_030 |
0.1322166179 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_004158_050 |
0.1339547592 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 9 |
Hb_001021_210 |
0.1363611316 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002151_010 |
0.1383742153 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 11 |
Hb_000836_350 |
0.1389638283 |
- |
- |
PREDICTED: uncharacterized protein LOC105642938 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001571_030 |
0.1403324848 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X1 [Jatropha curcas] |
| 13 |
Hb_010620_050 |
0.1415279127 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 14 |
Hb_000529_130 |
0.1423680076 |
- |
- |
hypothetical protein POPTR_0007s14320g [Populus trichocarpa] |
| 15 |
Hb_000562_070 |
0.1451526825 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001582_020 |
0.1486466185 |
- |
- |
plastid acyl-ACP thioesterase [Vernicia fordii] |
| 17 |
Hb_002686_300 |
0.1500610399 |
- |
- |
adenosine diphosphatase, putative [Ricinus communis] |
| 18 |
Hb_003687_080 |
0.1505468087 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 19 |
Hb_018133_100 |
0.1511064423 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000968_060 |
0.1516361561 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |