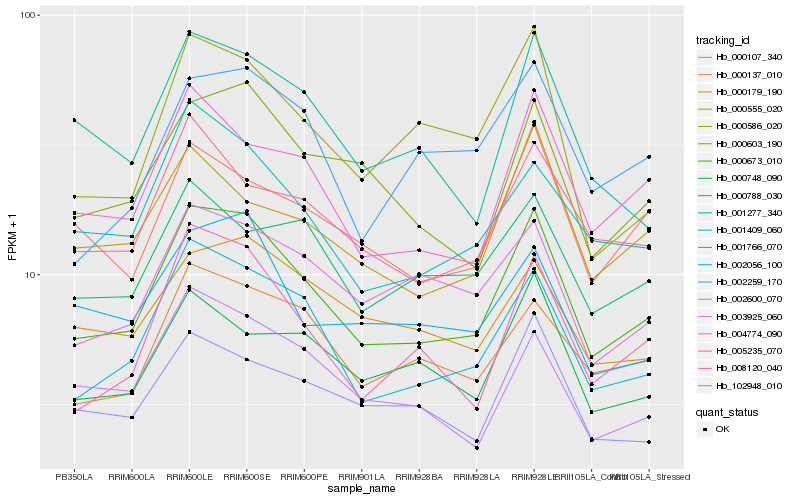
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000673_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636554 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001766_070 |
0.0649987321 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008120_040 |
0.0863758821 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 4 |
Hb_005235_070 |
0.0875634556 |
- |
- |
PREDICTED: plastid lipid-associated protein 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000137_010 |
0.0925890096 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 6 |
Hb_000603_190 |
0.0931559525 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 7 |
Hb_001409_060 |
0.0948564891 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 8 |
Hb_102948_010 |
0.0970481814 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000107_340 |
0.0988930078 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105637652 [Jatropha curcas] |
| 10 |
Hb_003925_060 |
0.1001938222 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002600_070 |
0.1012991135 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 12 |
Hb_004774_090 |
0.1022567084 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 13 |
Hb_000555_020 |
0.1042328072 |
- |
- |
APO protein 2, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000748_090 |
0.1050838148 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 15 |
Hb_000586_020 |
0.1051221861 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein F383_02497 [Gossypium arboreum] |
| 16 |
Hb_000179_190 |
0.1053521311 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 17 |
Hb_001277_340 |
0.105730846 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 18 |
Hb_002056_100 |
0.1061819981 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 19 |
Hb_000788_030 |
0.1062754847 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 20 |
Hb_002259_170 |
0.1062771044 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |