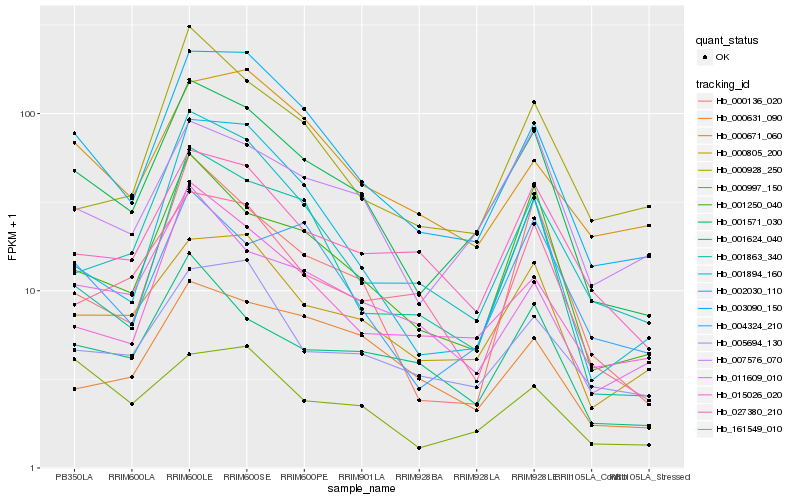
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001571_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003090_150 |
0.1067116212 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 3 |
Hb_001250_040 |
0.1123873005 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 4 |
Hb_000805_200 |
0.112966837 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_007576_070 |
0.123260565 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X3 [Jatropha curcas] |
| 6 |
Hb_027380_210 |
0.1365535075 |
- |
- |
PREDICTED: uncharacterized protein LOC105634030 [Jatropha curcas] |
| 7 |
Hb_000136_020 |
0.1391506907 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g80440-like [Jatropha curcas] |
| 8 |
Hb_004324_210 |
0.1403324848 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 9 |
Hb_001624_040 |
0.1419372188 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_015026_020 |
0.1432036468 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 11 |
Hb_001894_160 |
0.1438041312 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005694_130 |
0.1450960673 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 13 |
Hb_001863_340 |
0.1458269627 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 14 |
Hb_011609_010 |
0.1485947301 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 15 |
Hb_161549_010 |
0.1492090581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000928_250 |
0.1536005618 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 17 |
Hb_000997_150 |
0.1559457545 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000631_090 |
0.1595014534 |
- |
- |
PREDICTED: uncharacterized protein LOC105640619 [Jatropha curcas] |
| 19 |
Hb_002030_110 |
0.159880662 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 20 |
Hb_000671_060 |
0.1603880402 |
- |
- |
protein binding protein, putative [Ricinus communis] |