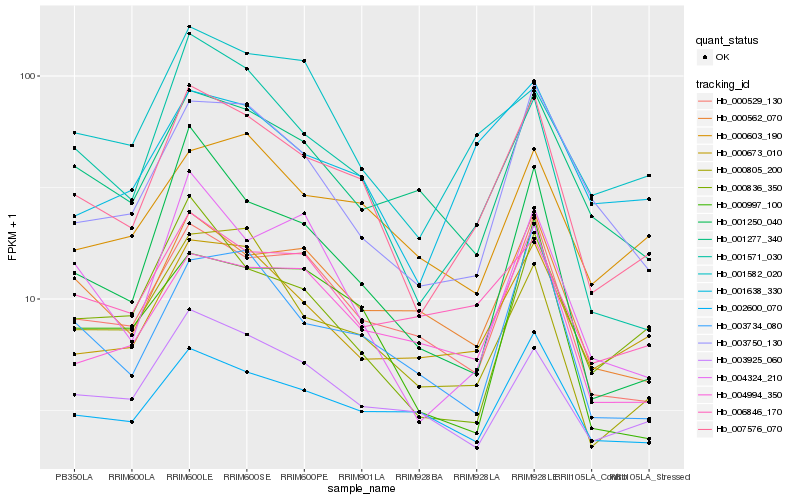
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007576_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X3 [Jatropha curcas] |
| 2 |
Hb_000805_200 |
0.1150068439 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000529_130 |
0.117231017 |
- |
- |
hypothetical protein POPTR_0007s14320g [Populus trichocarpa] |
| 4 |
Hb_001571_030 |
0.123260565 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003734_080 |
0.1237122597 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 6 |
Hb_000673_010 |
0.1245222905 |
- |
- |
PREDICTED: uncharacterized protein LOC105636554 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001638_330 |
0.1285428015 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000562_070 |
0.1296321721 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001250_040 |
0.1300706768 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 10 |
Hb_004994_350 |
0.1304260161 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004324_210 |
0.1317737817 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 12 |
Hb_003925_060 |
0.1318999407 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003750_130 |
0.13317586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001582_020 |
0.1340239634 |
- |
- |
plastid acyl-ACP thioesterase [Vernicia fordii] |
| 15 |
Hb_000603_190 |
0.134639117 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 16 |
Hb_000997_100 |
0.1356524548 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000836_350 |
0.1365310328 |
- |
- |
PREDICTED: uncharacterized protein LOC105642938 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001277_340 |
0.1385814356 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 19 |
Hb_006846_170 |
0.1388671872 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 20 |
Hb_002600_070 |
0.1404034687 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |