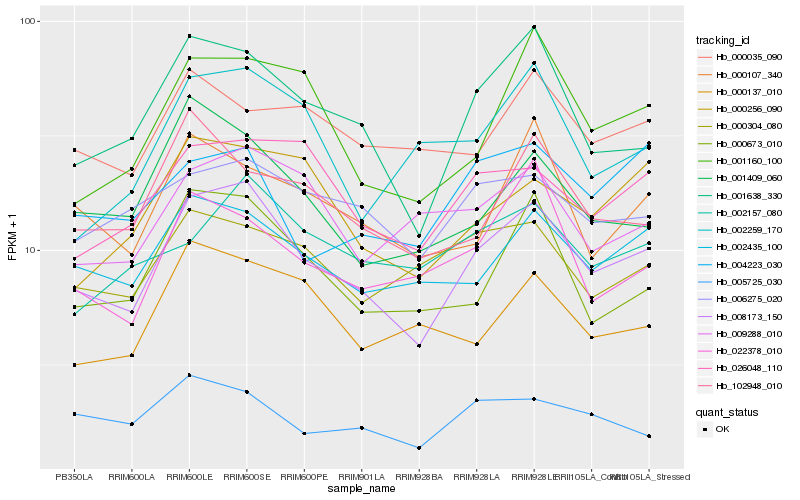
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008173_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645812 [Jatropha curcas] |
| 2 |
Hb_001638_330 |
0.1002591473 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002435_100 |
0.1121516228 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF217 [Jatropha curcas] |
| 4 |
Hb_000673_010 |
0.1135241971 |
- |
- |
PREDICTED: uncharacterized protein LOC105636554 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001160_100 |
0.1231721324 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 6, chloroplastic [Jatropha curcas] |
| 6 |
Hb_026048_110 |
0.1242168557 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 7 |
Hb_001409_060 |
0.1254037131 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 8 |
Hb_006275_020 |
0.1274567279 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 9 |
Hb_000256_090 |
0.1280443891 |
- |
- |
PREDICTED: uncharacterized protein LOC105637579 [Jatropha curcas] |
| 10 |
Hb_000107_340 |
0.1285497514 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105637652 [Jatropha curcas] |
| 11 |
Hb_009288_010 |
0.1303317706 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_102948_010 |
0.1329260271 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000137_010 |
0.1337897095 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 14 |
Hb_005725_030 |
0.1343017179 |
- |
- |
PREDICTED: U-box domain-containing protein 27-like [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_000304_080 |
0.1351067757 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002157_080 |
0.1353368283 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 17 |
Hb_022378_010 |
0.1357092051 |
- |
- |
PREDICTED: uncharacterized protein LOC105634715 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002259_170 |
0.1376057283 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_004223_030 |
0.1400510476 |
- |
- |
PREDICTED: uncharacterized protein LOC105635347 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000035_090 |
0.1402123665 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |