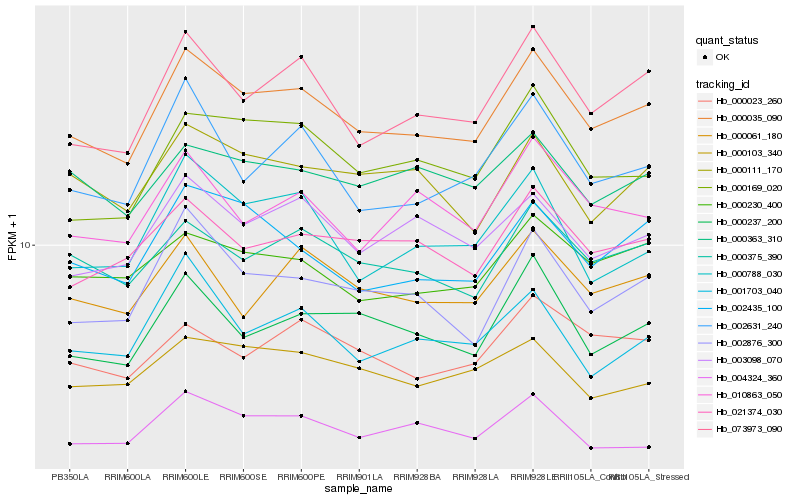
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_090 |
0.0 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 2 |
Hb_073973_090 |
0.0579488889 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 3 |
Hb_000375_390 |
0.0645379907 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 4 |
Hb_002876_300 |
0.067191656 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_002435_100 |
0.0707731548 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF217 [Jatropha curcas] |
| 6 |
Hb_003098_070 |
0.0714066474 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 7 |
Hb_000169_020 |
0.0719476721 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 8 |
Hb_021374_030 |
0.0723359884 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 9 |
Hb_000237_200 |
0.0731214717 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 10 |
Hb_000230_400 |
0.0734762756 |
- |
- |
PREDICTED: protein ABCI12, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_002631_240 |
0.0749901117 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 12 |
Hb_000061_180 |
0.0751081305 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 13 |
Hb_000111_170 |
0.077279102 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000788_030 |
0.0778909643 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 15 |
Hb_010863_050 |
0.0799085952 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 16 |
Hb_004324_360 |
0.0802433651 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000103_340 |
0.0807088613 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 18 |
Hb_000023_260 |
0.0807484038 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000363_310 |
0.0808745384 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 20 |
Hb_001703_040 |
0.0811892336 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |