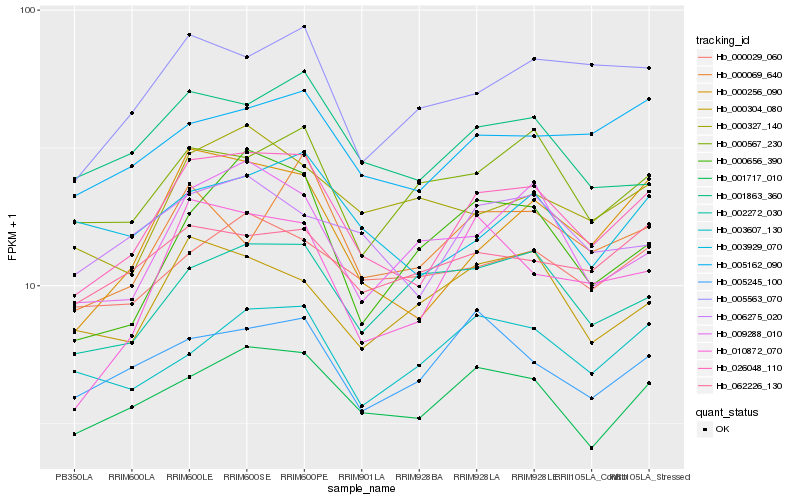
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026048_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 2 |
Hb_000256_090 |
0.0765517246 |
- |
- |
PREDICTED: uncharacterized protein LOC105637579 [Jatropha curcas] |
| 3 |
Hb_001717_010 |
0.0801952917 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 4 |
Hb_005162_090 |
0.0913854357 |
- |
- |
PREDICTED: uncharacterized protein LOC105648425 [Jatropha curcas] |
| 5 |
Hb_010872_070 |
0.0965156113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005563_070 |
0.0968545388 |
- |
- |
PREDICTED: uncharacterized protein LOC105630108 [Jatropha curcas] |
| 7 |
Hb_006275_020 |
0.0969797091 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 8 |
Hb_001863_360 |
0.0998054268 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 9 |
Hb_009288_010 |
0.1009601122 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_000029_060 |
0.1015874448 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 11 |
Hb_000069_640 |
0.1033434699 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 12 |
Hb_005245_100 |
0.1038438257 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_062226_130 |
0.105386625 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 14 |
Hb_000567_230 |
0.1060116293 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 15 |
Hb_003929_070 |
0.1066193946 |
- |
- |
CBL-interacting serine/threonine-protein kinase 1 isoform 3 [Theobroma cacao] |
| 16 |
Hb_002272_030 |
0.1075266981 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 17 |
Hb_000304_080 |
0.1075386955 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000327_140 |
0.1075629045 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000656_390 |
0.107966966 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003607_130 |
0.1088453489 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |