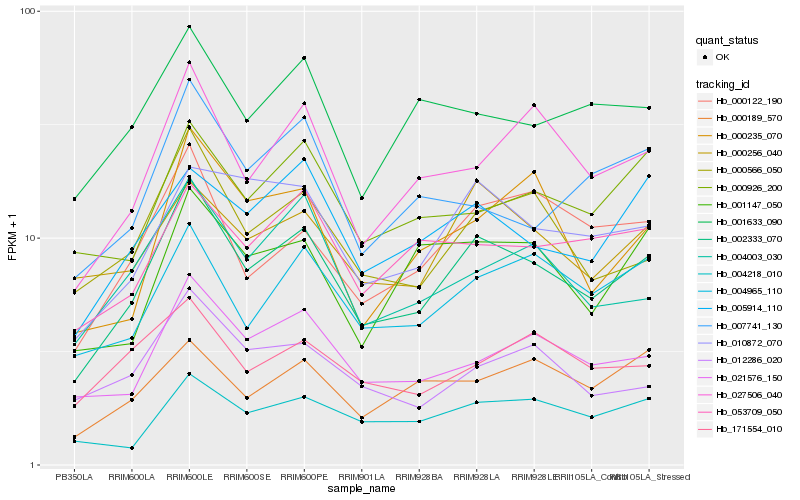
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002333_070 |
0.0 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4-like [Jatropha curcas] |
| 2 |
Hb_000566_050 |
0.1054381271 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 3 |
Hb_021576_150 |
0.1084515556 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 4 |
Hb_027506_040 |
0.1105609182 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000122_190 |
0.1143532703 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_010872_070 |
0.1145930649 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004965_110 |
0.1181247958 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 8 |
Hb_005914_110 |
0.1192887862 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 9 |
Hb_004003_030 |
0.1208834543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000256_040 |
0.1227240367 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_004218_010 |
0.1236598946 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 12 |
Hb_053709_050 |
0.125172248 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_001633_090 |
0.1252087097 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 14 |
Hb_012286_020 |
0.1283122329 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 15 |
Hb_001147_050 |
0.1298144908 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000926_200 |
0.1304033479 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_000235_070 |
0.1316135219 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 18 |
Hb_171554_010 |
0.1326275122 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 19 |
Hb_000189_570 |
0.1332585712 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 20 |
Hb_007741_130 |
0.134407655 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |