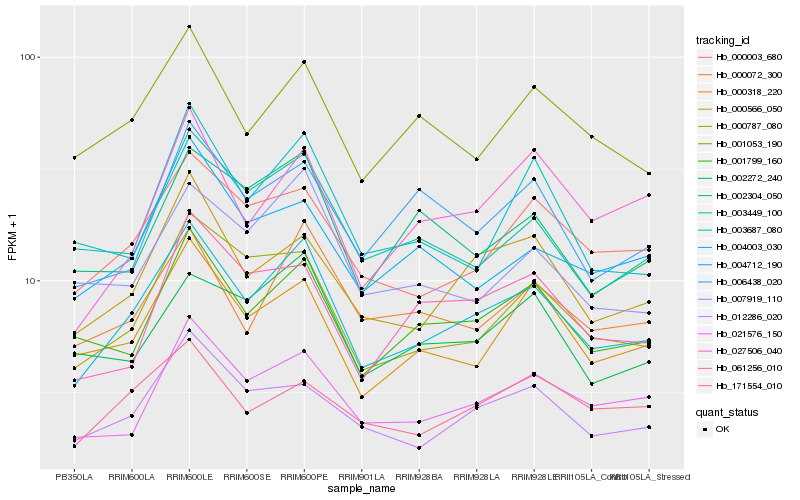
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004003_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000566_050 |
0.0917391787 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 3 |
Hb_001799_160 |
0.0946480837 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_000787_080 |
0.0948953123 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 5 |
Hb_000003_680 |
0.0961295819 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 6 |
Hb_021576_150 |
0.097366265 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 7 |
Hb_000318_220 |
0.1000864355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_012286_020 |
0.1004811756 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 9 |
Hb_001053_190 |
0.1030830173 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 10 |
Hb_000072_300 |
0.1041886556 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 11 |
Hb_007919_110 |
0.1104292944 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 12 |
Hb_002272_240 |
0.1121621411 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_003687_080 |
0.113155852 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 14 |
Hb_004712_190 |
0.1140301442 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 15 |
Hb_003449_100 |
0.1155244224 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 16 |
Hb_002304_050 |
0.1167965425 |
- |
- |
pyruvate kinase, putative [Ricinus communis] |
| 17 |
Hb_027506_040 |
0.116812239 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_171554_010 |
0.117574348 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 19 |
Hb_006438_020 |
0.117578438 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 20 |
Hb_061256_010 |
0.1180323366 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |