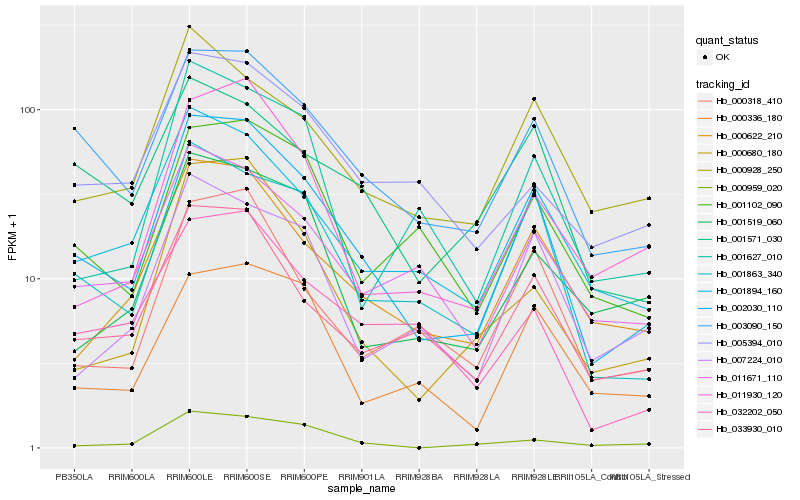
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002030_110 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL32-like [Jatropha curcas] |
| 2 |
Hb_003090_150 |
0.1112174068 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 3 |
Hb_001863_340 |
0.1233586991 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 4 |
Hb_001894_160 |
0.1287514523 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000622_210 |
0.1304737129 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_033930_010 |
0.1351845549 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 7 |
Hb_000680_180 |
0.1434453492 |
- |
- |
hypothetical protein JCGZ_15248 [Jatropha curcas] |
| 8 |
Hb_011671_110 |
0.1480748908 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 9 |
Hb_000318_410 |
0.148775523 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 10 |
Hb_032202_050 |
0.149635153 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_011930_120 |
0.1522201344 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001102_090 |
0.1536817629 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 13 |
Hb_000336_180 |
0.1581044034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007224_010 |
0.158875239 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 15 |
Hb_001519_060 |
0.1598783786 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_001571_030 |
0.159880662 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000928_250 |
0.1605324048 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_001627_010 |
0.1617230414 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 19 |
Hb_005394_010 |
0.1617642512 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 20 |
Hb_000959_020 |
0.1623575124 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |