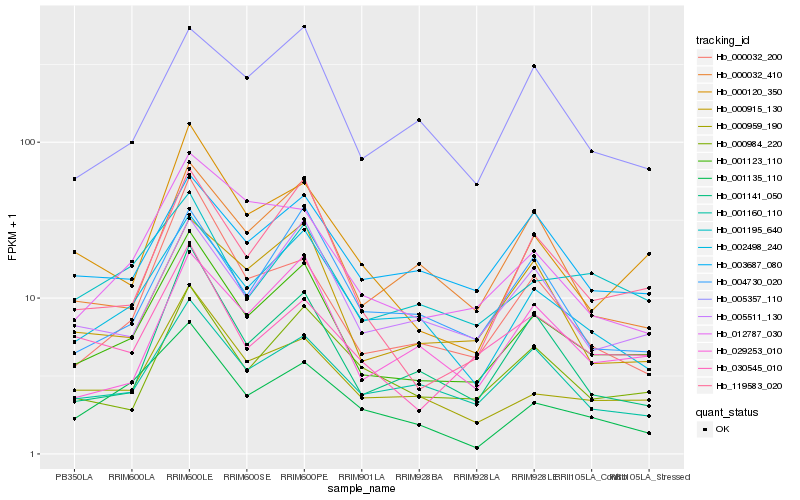
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_119583_020 |
0.1094077155 |
- |
- |
PREDICTED: uncharacterized protein LOC105628090 [Jatropha curcas] |
| 3 |
Hb_000959_190 |
0.1141449069 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 4 |
Hb_002498_240 |
0.1147320817 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005511_130 |
0.1268313116 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 6 |
Hb_000984_220 |
0.1269582324 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 7 |
Hb_001160_110 |
0.1289327995 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 8 |
Hb_000120_350 |
0.1290760914 |
- |
- |
PREDICTED: uncharacterized protein LOC104422872 isoform X1 [Eucalyptus grandis] |
| 9 |
Hb_029253_010 |
0.1336075078 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 10 |
Hb_001141_050 |
0.134486787 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001195_640 |
0.1349930333 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 12 |
Hb_000915_130 |
0.1354776991 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 13 |
Hb_030545_010 |
0.1381493794 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 14 |
Hb_003687_080 |
0.1400410168 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 15 |
Hb_001135_110 |
0.1406293417 |
- |
- |
- |
| 16 |
Hb_012787_030 |
0.141007148 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 17 |
Hb_000032_410 |
0.1418346878 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 18 |
Hb_000032_200 |
0.1426309682 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 19 |
Hb_005357_110 |
0.1451371967 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 20 |
Hb_004730_020 |
0.1452021238 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |