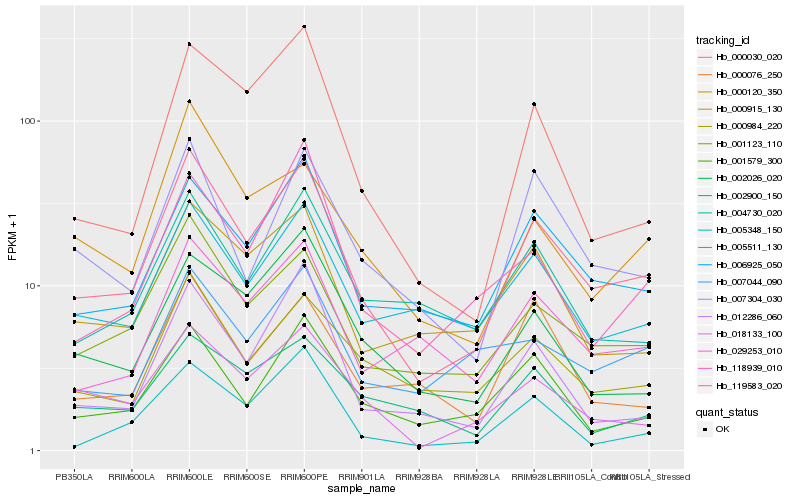
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_119583_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628090 [Jatropha curcas] |
| 2 |
Hb_001123_110 |
0.1094077155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006925_050 |
0.1173164589 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 4 |
Hb_018133_100 |
0.133665076 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000984_220 |
0.1400556555 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 6 |
Hb_001579_300 |
0.1415048554 |
- |
- |
PREDICTED: probable polygalacturonase At1g80170 [Jatropha curcas] |
| 7 |
Hb_007304_030 |
0.1416399125 |
- |
- |
PREDICTED: uncharacterized protein LOC105635926 isoform X2 [Jatropha curcas] |
| 8 |
Hb_029253_010 |
0.1438614977 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 9 |
Hb_005511_130 |
0.1438668102 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 10 |
Hb_000915_130 |
0.1453584596 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 11 |
Hb_004730_020 |
0.1515209972 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 12 |
Hb_000030_020 |
0.1517394107 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 13 |
Hb_012286_060 |
0.1535919206 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_007044_090 |
0.1547000244 |
- |
- |
PREDICTED: isoprenylcysteine alpha-carbonyl methylesterase ICME isoform X2 [Jatropha curcas] |
| 15 |
Hb_000120_350 |
0.1547446738 |
- |
- |
PREDICTED: uncharacterized protein LOC104422872 isoform X1 [Eucalyptus grandis] |
| 16 |
Hb_002026_020 |
0.1551703236 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 17 |
Hb_118939_010 |
0.1554694429 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_005348_150 |
0.1588833876 |
- |
- |
PREDICTED: actin-100-like [Jatropha curcas] |
| 19 |
Hb_000076_250 |
0.1622247901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002900_150 |
0.163205964 |
- |
- |
PREDICTED: syntaxin-81 [Jatropha curcas] |