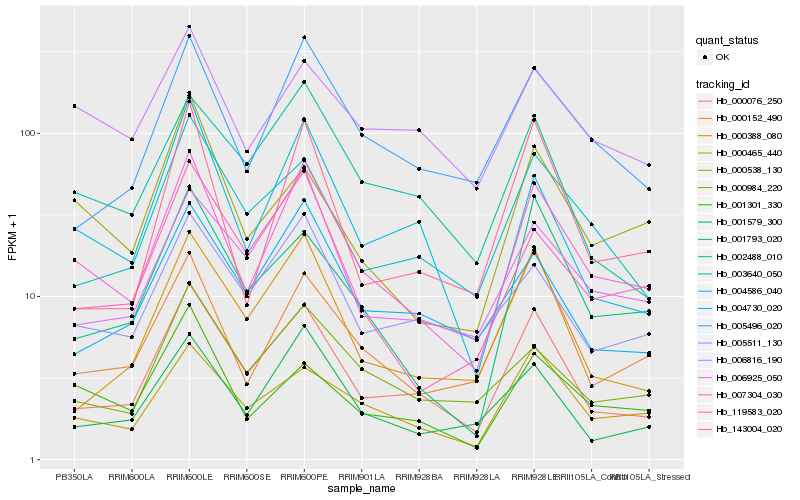
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007304_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635926 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000076_250 |
0.1189192098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000465_440 |
0.1332995454 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2-like [Jatropha curcas] |
| 4 |
Hb_005496_020 |
0.1333072388 |
- |
- |
PREDICTED: uncharacterized protein LOC105630867 [Jatropha curcas] |
| 5 |
Hb_001579_300 |
0.1352580481 |
- |
- |
PREDICTED: probable polygalacturonase At1g80170 [Jatropha curcas] |
| 6 |
Hb_119583_020 |
0.1416399125 |
- |
- |
PREDICTED: uncharacterized protein LOC105628090 [Jatropha curcas] |
| 7 |
Hb_000152_490 |
0.1442146497 |
- |
- |
PREDICTED: (6-4)DNA photolyase [Jatropha curcas] |
| 8 |
Hb_006925_050 |
0.1449911182 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 9 |
Hb_001301_330 |
0.1450958902 |
- |
- |
OTU-like cysteine protease family protein [Populus trichocarpa] |
| 10 |
Hb_000984_220 |
0.1487363683 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 11 |
Hb_005511_130 |
0.1490909222 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 12 |
Hb_004586_040 |
0.1503496989 |
transcription factor |
TF Family: MIKC |
K-box region and MADS-box transcription factor family protein [Theobroma cacao] |
| 13 |
Hb_006816_190 |
0.1506454801 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 14 |
Hb_001793_020 |
0.1527921225 |
- |
- |
PREDICTED: uncharacterized protein LOC105628624 [Jatropha curcas] |
| 15 |
Hb_003640_050 |
0.155980756 |
- |
- |
PREDICTED: protein LUTEIN DEFICIENT 5, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_000538_130 |
0.1560639508 |
- |
- |
phytoene synthase 2 [Manihot esculenta] |
| 17 |
Hb_004730_020 |
0.1561844317 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 18 |
Hb_143004_020 |
0.1581739675 |
- |
- |
PREDICTED: phosphopantothenate--cysteine ligase 2-like [Jatropha curcas] |
| 19 |
Hb_002488_010 |
0.1599675203 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 20 |
Hb_000388_080 |
0.161218713 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |