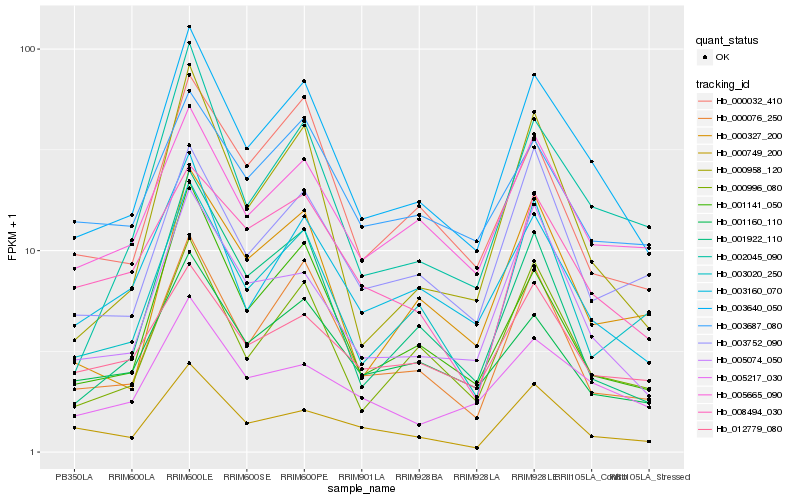
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003640_050 |
0.0 |
- |
- |
PREDICTED: protein LUTEIN DEFICIENT 5, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_000076_250 |
0.1029014287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000996_080 |
0.103471312 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 4 |
Hb_005665_090 |
0.1140199389 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 5 |
Hb_005074_050 |
0.1142560842 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 6 |
Hb_003160_070 |
0.1157864636 |
- |
- |
PREDICTED: VIN3-like protein 1 [Jatropha curcas] |
| 7 |
Hb_001141_050 |
0.1162554207 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002045_090 |
0.1205488834 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 9 |
Hb_001160_110 |
0.1207464858 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 10 |
Hb_000032_410 |
0.1232312719 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 11 |
Hb_000327_200 |
0.1245513491 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 12 |
Hb_000958_120 |
0.1251439889 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 13 |
Hb_005217_030 |
0.1267949549 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 14 |
Hb_003752_090 |
0.1275293887 |
- |
- |
chitinase, putative [Ricinus communis] |
| 15 |
Hb_003020_250 |
0.1321292789 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 16 |
Hb_001922_110 |
0.1334693948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003687_080 |
0.1344928907 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 18 |
Hb_008494_030 |
0.1351574938 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_012779_080 |
0.1353594774 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000749_200 |
0.1363078296 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |