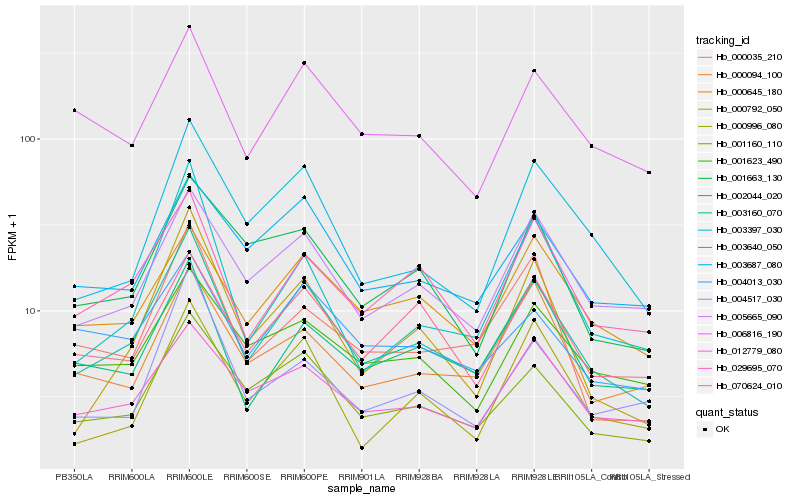
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003160_070 |
0.0 |
- |
- |
PREDICTED: VIN3-like protein 1 [Jatropha curcas] |
| 2 |
Hb_001160_110 |
0.0843552565 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 3 |
Hb_005665_090 |
0.1075914267 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 4 |
Hb_029695_070 |
0.1087888965 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_003640_050 |
0.1157864636 |
- |
- |
PREDICTED: protein LUTEIN DEFICIENT 5, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_012779_080 |
0.1178729569 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 7 |
Hb_000792_050 |
0.1213330747 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_003687_080 |
0.1221122963 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 9 |
Hb_000645_180 |
0.122420992 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 10 |
Hb_006816_190 |
0.1228685497 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 11 |
Hb_002044_020 |
0.123577748 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 12 |
Hb_070624_010 |
0.1264898135 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 13 |
Hb_001623_490 |
0.1267550928 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001663_130 |
0.1270930222 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 15 |
Hb_003397_030 |
0.127460372 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004013_030 |
0.1286876123 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 17 |
Hb_000094_100 |
0.1305319465 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 18 |
Hb_004517_030 |
0.1305552979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000035_210 |
0.1316520419 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 20 |
Hb_000996_080 |
0.1319413802 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |