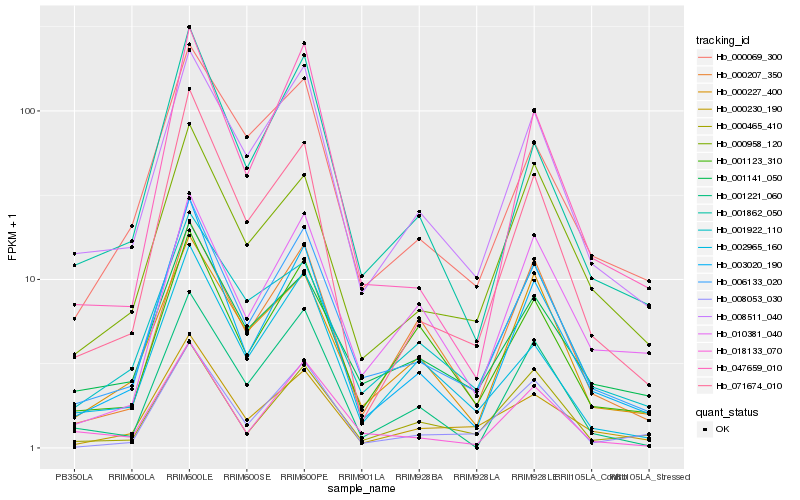
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006133_020 |
0.0 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 2 |
Hb_008511_040 |
0.0728933167 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 3 |
Hb_003020_190 |
0.0953446279 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 4 |
Hb_001862_050 |
0.1100812892 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 5 |
Hb_000227_400 |
0.1109506783 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 6 |
Hb_001141_050 |
0.1112500753 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000230_190 |
0.1114773815 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001123_310 |
0.112476573 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 9 |
Hb_018133_070 |
0.1133220212 |
- |
- |
PREDICTED: RING-H2 finger protein ATL43 [Jatropha curcas] |
| 10 |
Hb_000069_300 |
0.1158599322 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 11 |
Hb_008053_030 |
0.1165165198 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 12 |
Hb_001922_110 |
0.1171205353 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000958_120 |
0.1194793767 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 14 |
Hb_047659_010 |
0.1209766261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000207_350 |
0.1211524684 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 16 |
Hb_071674_010 |
0.1221124434 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 17 |
Hb_000465_410 |
0.1248655288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010381_040 |
0.1258909578 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 19 |
Hb_001221_060 |
0.1265026607 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 20 |
Hb_002965_160 |
0.1272432719 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |