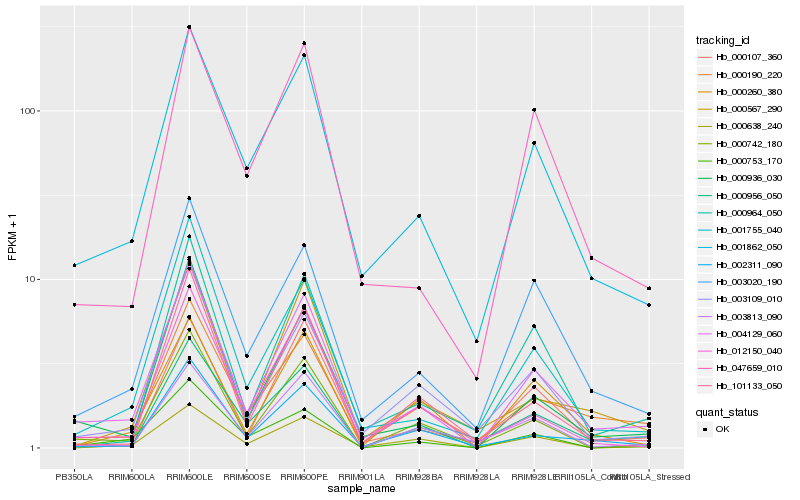
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004129_060 |
0.0 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 2 |
Hb_001862_050 |
0.1033136979 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 3 |
Hb_000936_030 |
0.1120112243 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 4 |
Hb_003020_190 |
0.1224199857 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 5 |
Hb_000190_220 |
0.1269542616 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 6 |
Hb_001755_040 |
0.1316680323 |
- |
- |
PREDICTED: cyclin-A2-4-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_003109_010 |
0.1339153696 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 8 |
Hb_000742_180 |
0.1369850704 |
- |
- |
hypothetical protein POPTR_0011s16640g [Populus trichocarpa] |
| 9 |
Hb_047659_010 |
0.1422711833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002311_090 |
0.143956585 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 11 |
Hb_012150_040 |
0.1448190744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000956_050 |
0.1458149987 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000753_170 |
0.1484311003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000638_240 |
0.1484514028 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Prunus mume] |
| 15 |
Hb_000107_360 |
0.1485605985 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 16 |
Hb_000567_290 |
0.1502554505 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 17 |
Hb_000964_050 |
0.1511987801 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 18 |
Hb_003813_090 |
0.15131054 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 19 |
Hb_101133_050 |
0.1515392888 |
- |
- |
PREDICTED: condensin complex subunit 3 [Jatropha curcas] |
| 20 |
Hb_000260_380 |
0.1518355395 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |