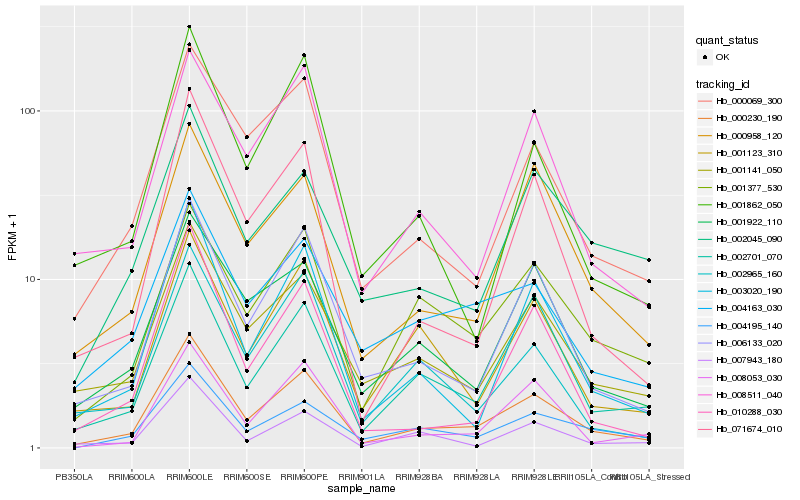
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_190 |
0.0 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006133_020 |
0.1114773815 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 3 |
Hb_001141_050 |
0.1172135184 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_003020_190 |
0.1216416448 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 5 |
Hb_001377_530 |
0.1249214687 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 6 |
Hb_000958_120 |
0.1268885714 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 7 |
Hb_000069_300 |
0.1273952113 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 8 |
Hb_004195_140 |
0.1304018858 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 9 |
Hb_004163_030 |
0.1324585603 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 10 |
Hb_001922_110 |
0.132728966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_008511_040 |
0.1352369631 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 12 |
Hb_071674_010 |
0.1357724556 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 13 |
Hb_001123_310 |
0.136773437 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 14 |
Hb_002701_070 |
0.1371429857 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 15 |
Hb_008053_030 |
0.1383392996 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 16 |
Hb_002965_160 |
0.1458393196 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 17 |
Hb_007943_180 |
0.1488370364 |
- |
- |
PREDICTED: uncharacterized protein LOC105637734 [Jatropha curcas] |
| 18 |
Hb_010288_030 |
0.1491766104 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 19 |
Hb_002045_090 |
0.1501006173 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 20 |
Hb_001862_050 |
0.1527292792 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |